














Genetics a marvel science has become an indispensable component of almost all research in modern biology and medicine. Almost all research publications investigating any biological process, from the molecular level all the way to the population level, rely on the “genetic approach” to gain understanding of that process. Thus, no student of the life sciences can afford to be ignorant of the science of genetics.
Genetically Modified types of plants, animals, and microbes have been developed for human foods, drugs, and myriad other uses. Molecular genetics is the central foundation of the burgeoning biotechnology industry. At the philosophical level, genetics has presented humans with a large number of ethical dilemmas, which regularly surface in the media. Some examples are genetically modified foods, eugenics, privacy of genetic information about individuals, and loss of genetic diversity in nature. Students must be knowledgeable about genetics in order to understand these issues and make informed decisions about them. Lastly, genetic insight has radically affected the human worldview—the way we see ourselves in relation to other organisms.
This is book has been years of efforts, upgrades and perfection. We proudly present to you the Inheritance book, which is a compiled version of all genetic concepts.
Each chapter begins with the “snapshots” which is overview of the chapter. This is aimed to give the students what it is going to be in next leaves and also for the end moment preparations.
Most chapters have a solved problems for students to understand the approach. This exercise grew from the idea that a genetics problem represents only the tip of a vast iceberg of knowledge. It is only when the underlying levels of knowledge are exposed that the problem can be solved in a constructive manner.
The critical thinking questions at the end of every chapter, aimed to be the brain teasers for students, invoking thinking and analysis.
The problems at the end of the book are prefaced by Solved Problems that illustrate the ways that geneticists apply principles to experimental data. Research in science education has shown that this application of principles is a process that professionals find second nature, whereas students find it a major stumbling block.
Throughout the chapters, boxed Messages provide interesting and valuable facts of genetics and biology.
We provide the web link at
Students are advised to use the web source for PPTs and lecture videos of the chapters. The web source also provides over 2000 questions on the Unit.
Thus we the Academic Team at Biotecnika proudly present to you to the second edition of Study Material.

MENDELISM
CONCEPTS
8.3 EXTENSIONS

8.4 GENE MAPPING
8.4.1
EXTRA CHROMOSOMAL
MICROBIAL GENETICS

8.7 HUMAN GENETICS
Linked
QUANTITATIVE GENETICS:

1. Mendel was the first person who attempted to understand the basics of genetics in systematic way

2. He choose Garden Pea Plant, for his studies and came up with some interesting facts
3. He said, there is some important factor in plants, which carried the characters through generations.
4. He observed, that some characters appear most of the time in plant progeny compared to others, he termed them as Dominant characters
5. Some characters were very rarely observed and he termed them as
6. He chose 7 different characters in pea plant for his studies
7. He conducted several crosses of these plants and came up with the 2 most important and basic laws of inheritance
8. Law of segregation: 2 alleles of a gene always segregate during gamete formation.
9. Law of Independent assortment: The alleles segregate independent of each other during gamete formation
10. Monohybrid cross: Observing the inheritance pattern with only one gene into consideration Monohy-

brid ratio is 3:1
11. Dihybrid cross
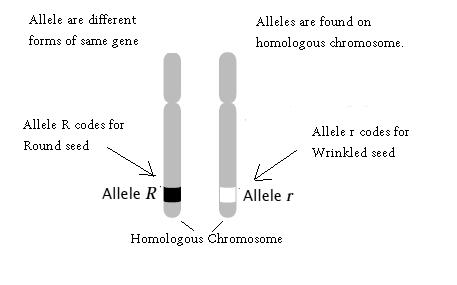
12. Concept of Allele: Alleles are the different forms of same gene. They have similar sequences and code for similar enzymes, which may/may not carry out same biochemical reaction. Allele occupies same location on homologous chromosomes.
13. Dominant allele has very strong promoters and other genetic elements, which makes it to get expressed most of the time in progeny and it carries out a prominent biochemical reaction, hence becoming dominant gene
14. Whereas the recessive allele, has weak promotes and other suppressing genetic elements, which makes it not to get expressed more often and thus its enzyme does not catalyze any prominent reaction, making it to be recessive
15. But the Mendelian pattern of inheritance does not work all the time
16. In many species, even in Pea plant itself, we observe many deviations from his laws of Inheritance. The main reason being existence of Linkage, Crossing over, Expressivity and Penetrance, Cytoplasmic inheritance, Gene interactions, maternal effect…etc.
October 19, 1989, cool breeze touched Mr. Pandu’s legs. He was waiting outside a hospital, where his pregnant wife was about to deliver his first child. His relatives were eager to welcome the child to the world. When the child was delivered, they were very happy, but no one was surprised!! How did they know that a human baby is coming? Why they didn’t expect a lion or a mango tree?
Ever since the human started thinking about the life, he always puzzled about how we resemble our parents. How a Mango tree gives seeds of Mango tree? How each organism is unique, yet reproduces a similar organism. There is something or some factors in us, which is making us to behave like humans and reproduce a human baby.
Way back in 1850s, a priest Gregor Mendel was interested in plant breeding and maintained a pea plant farm in his backyard. He observed some peculiar characters in pea plants and noted them down. He also did some controlled experiments just by curiosity. The observations and explanation he gave are the ground breaking in field of biology. It was the baby step by human in the world of Genetics. He has laid down some important rules by which pea plants pass on their characters to the offspring. In this section we shall learn about Mendel
Gene is a sequence of enzyme. That enzyme will catalyze a particular biochemical reaction and we get the product of the reaction. The gene is just a sequence, which we cannot see.
The external appearance is called as In human, phenotype can be hair color, body color, digestive capacity, food habits...etc; whereas phenotypes in microbial world is measured in terms on culture characteristics.

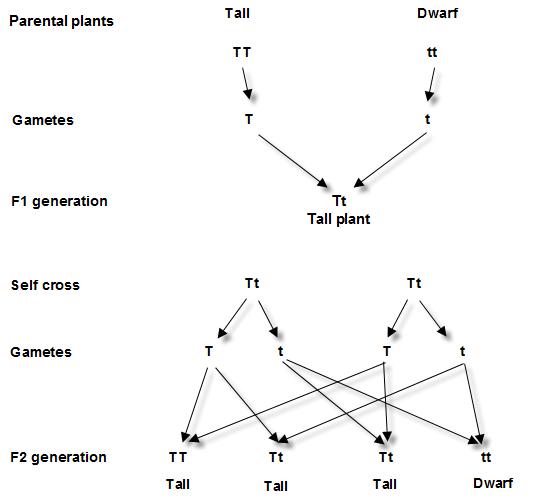
have is called as what the different genes you have are and what is their nature Mendel took 2 different plants, one is tall other is Dwarf and crossed them to obtain some progeny. What he observed was astonishing.
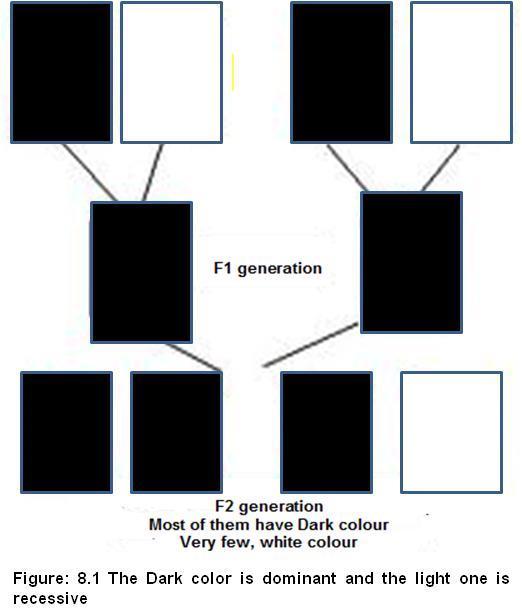
Most of the plants were Tall and very few were dwarf. That means Tall character is dominant over Dwarfness.
He said there is a dominant allele (Variant of same gene) “T” which makes plant tall and one more allele “t” which makes plant Dwarf. So any plant with “T” allele becomes Tall. Only when a plant gets 2 copies of ‘t” allele, it becomes Dwarf.
So we say any plant with “TT” “Tt” genotype : Dominant Tall plant;
Any plant with “tt” genotype: Recessive Dwarf plant. Above figure 8.1 shows the Dominant color and recessive color in a hypothetical cross. Why Mendel worked on pea plant, Pisum sativum for several years not any other?
Pea plants have short generation time, 3-4 months
Pea plants reproduce sexually, thus giving new combinations.
Pea plants produce many progeny at a time, easy for investigation in large scale
Pea plants have some very important characters with clear cut distinction.
Refer table 8.1. The 7 contrasting characters helped him to develop the laws of genetics.

Segregation means separation. What is separating? Alleles “T” and “t” come from your parents, one from father and the other from mother. Both the alleles sit together on homologous chromosomes, in your body cell and when gamete cells are getting formed during meiosis, those “T” and “t” separate from each other. This is called as

The homozygous plant with Tall character produces only T gamete.
It is called “ cause only 1 type of gamete “T” is produced
The other plant is dwarf in nature and it has “tt” characteristic. It is also called “Pure” breeding as it produces only 1 type of gamete ‘t”.
When these gametes were fused, we got a plant which i also Tall in nature, its having “Tt” genotype. It is called as Heterozygous generation
Now lets us go for 2
Figure8.2. Diagram illustrating the Mendel monohybrid cross. In 2nd generation,
We shall take 2 plants born to the same parental plants; we call it as “Tt” genotype.
Each plant can produce 2 type of gametes “T” and “t”. us see the above figure 8.2 and analyze what is happening.
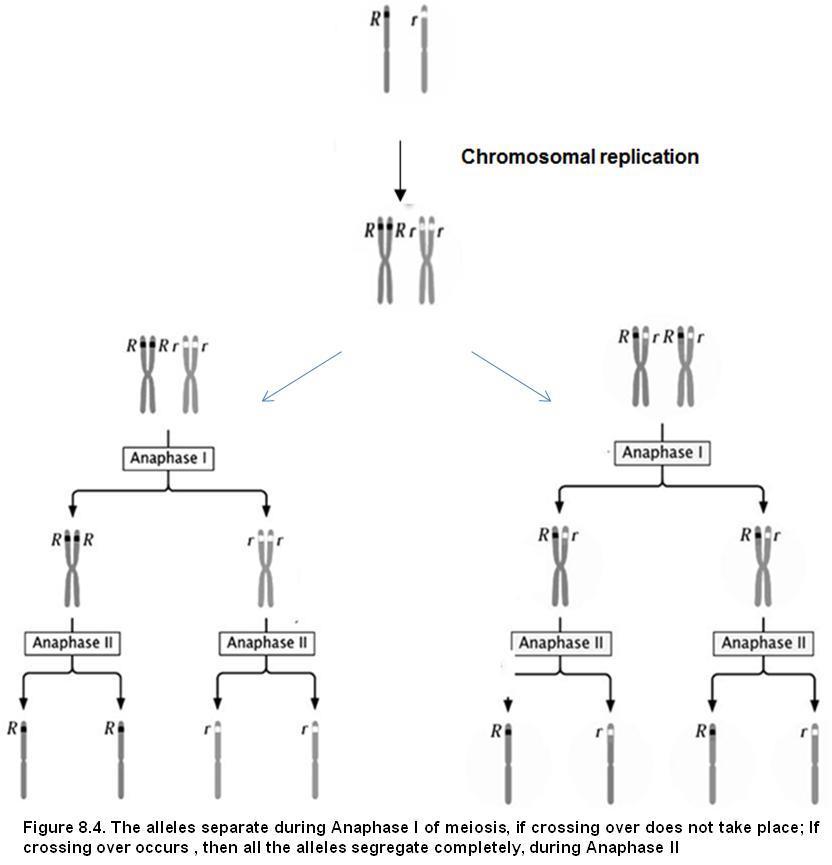
What you observe here? 3 plants are tall and 1 plant is dwarf. Hence we say, ration of Tall to Dwarf Monohybrid cross ratio Remember these are F2, Phenotypic ratio. We have seen that, during gamete formation the two alleles T and t, separated from each other. They move to different gametes. This concept is called Law of segregation. (Refer figure 8.3)
Note:
Figure8.3Separation of alleles in equalratio to 2newlyformedgamete cells-Segregation
1. The principle of segregation states that each individual organism possesses two alleles coding for a characteristic. These alleles segregate when gametes are formed, and one allele goes into each gamete.
2. The concept of dominance states that, when dominant and recessive alleles are present together, only the trait of the dominant allele is observed.
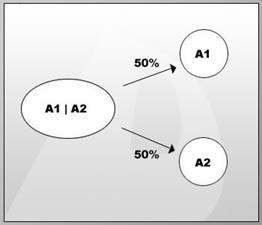

This is another rule of genetics, which occurs most of the time, but it is not a universal law. Let us understand what the meaning of it and how these alleles move independently. When we considered, 2 alleles of a gene, we found they separate during gamete formation. What if, we consider 2 genes at a time? Let us consider 2 different genes, R and Y. There are 2 alleles in each case, “R “r” and “Y” “y”. These genes code for different characters.
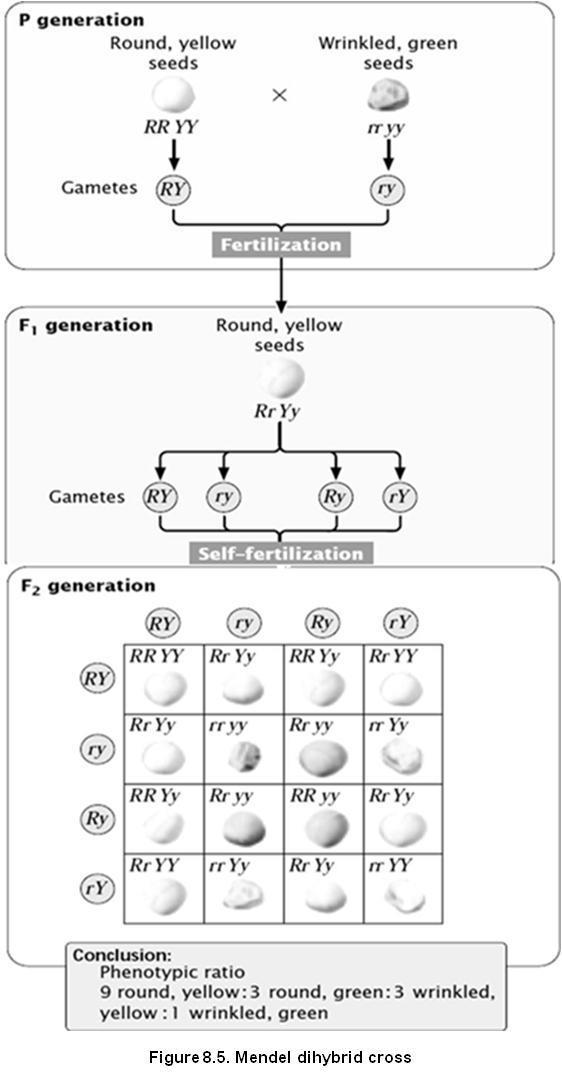
Let us take two pure breeding parent plants, RRYY and rryy. Lets us see what is happening in the below diagram 8.5.
In the following diagram, we have taken 2 plants and produced F2 plants.
We get 16 possible plants. In that we observe 9 plants are of one time, 3 are of another, rest 3 are of another and last 1 is a unique type.
You can see here, that 9 plants have dominant parental property (Round and yellow)
Whereas the last 1 plant has recessive parental property, and the rest 6 ( 3:3 ratio) plants are new types, differing from the parental
They are also called as recombinants
We need to be careful while assigning the phenotypes and also while producing the gametes. We need to pick only one of the allele forms each pair.
For example: if we have AABB then pick A and pick B, thus gamete is “AB”, if we have plant with AaBb, then we get 4 different gametes: AB, Ab, aB and ab.

As you can see in the adjacent diagram, the alleles on different loci are separated independently.
The allele R does not depend on the Y or y. It is completely independent of it.
Similarly allele, r, does not depend on Y or y, and vice versa.
Thus we can say that, the genes coding for Seed color and Seed shape are not linked and they are independent of each other
The principle of independent assortment states that genes coding for different characteristics separate independently of one another when gametes are formed located close together on the same chromosome do not assort independently Test cross:
In the above discussions so far, we have been stating homozygous plant, heterozygous plant…etc. But how did we come to know it’s genotype? Genotypes are not visible to our eyes. We can only see the phenotypes. Then how to find out the genotypes? We have a simple technique to find out the genotype of the unknown, the cross. Test cross is done to find out the genotype of unknown plant.
Test cross is any cross involving a plant with unknown genotype sive plant.
Ex: unknown genotype plant x rryy. It does not mean, recessive parent. It can be anyone with homozygous recessive condition. Back cross parent to the F1 progeny. In back crossing it is strictly the parent plant/animal. The backcross is useful in genetics studies for isolating (separating out) certain characteristics in a related group of animals or plants. In animal breeding, a backcross is often called a top cross. Grading usually refers to the mating of average, or “grade,” females to a superior male, then backcrossing the female offspring to the same or a similar sire.
Because of amutationand in-breeding,thetown of Sao Pedro,Brazilhasa 10%rate of twinkids


Genetics is a game played by genes. Genes flow from one generation to next, following their own rules and patterns. Thus it is very difficult to predict exactly which gene is going to which offspring. Even though you and your brother born to same parents, but yet you are different and we cannot predict what genes you have received and what your brother has got. Genetics also depends on chance factors. We have thousands of genes in entire genome. In that which gene is going to get express is completely a chance factor, so we use a mathematical technique called as Rule of prediction also called as Probability. This helps us to find out the chance of gene flow and chance of gaining the characters.
Consider a case of child birth. During one birth only 2 cases are possible either boy or girl child. Hence we say, out of 2 possibilities any one will occur. , in other words, ½ chance to get baby boy or ½ chance to get baby girl. This is always constant, never changes. For example in Plants, When Tall and Dwarf plants are crossed; we can get 3 Tall plants and 1 dwarf plant. Hence out of 4 plants, 3 will be Tall and 1 will be dwarf. Hence probability or chance to get Tall plant is ¾ and to get dwarf is ¼. This is always constant for all monohybrid crosses, never changes no matter what.
Additive rule

we can call
Either- Or
This rule is most used rule in genetics, used to find the probability of 2 independent events occurring at the same time
Ex: Chance that 2 boys occur in a family of 2 children. Here, probability of one boy – ½; Probability for second child to be boy = ½; Hence chance that both children are boys = ½ x ½ = ¼ Probability to get mentally challenged boy = 3/4 , for example; and chance of having a boy is ½ . Then chance of having mentally retarded boy = ¾ x ½ = 3/8
This is the most useful formula, which helps to predict the chance factors when things do not follow usual course. Remember this formula and can apply for most genetic problems. Although this formula looks little complicated, but when you solve some sample problems, you will be the best in it.
Here,
P – Probability

n – Total no. of events
x – no. of one possible event n-x – no. of other possible event
p - Probability of x event to occur
q – Probability of n-x event to occur.
! – Factorial; for example: 3! = 3x2x1 = 6 Is it possible to calculate the number of Gametes produced by the parents, if we know their genotypes? Yes it is possible, by understanding the rule of Probability. Imagine the parents “AA” and “aa” genotype. They can produce only one type fo gamete: “Aa”. Now if the parents have “Aa” and “aA” condition, then? We can have 4 possible children: AA, Aa, aA and aa. It is very simple to calculate these, what if we have multiple genes “AaBbCcDdEe” ? Let us see how to solve these. No.of gametes produced by this is given by formula: 2 “n” stands for no. of heterozygous pairs. In the above question, heterozygous pairs are: Aa, Bb, Cc, Dd and Ee. Hence n = 5 2n = 25 = 32. T
Now, let us move onto find out little complicated matter. If we have parents: AABBCcDdEe x AaBbCcDdEe
What is the chance that a child with “AABbccddEE” condition. Let us see how to solve it:
Thus total probability to get such a child is got by multiplying all these probabilities.
Humanbeingsshare7% of geneswithE.coli bacterium, 21%withworms, 90%withmiceand 98% withchimpanzees.

When a coin is tossed what is the probability that Head will appear?
Solution:
In a coin toss, head can come only once. Also, only 2 possible events, head or tail Therefore, Probability = 1 / 2 No need to use the formula
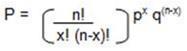
In afamily
Solution: What is Hence Whydid Productrulesays, multiplyindividual eventprobabilityvalue.Because,havingagirlschild completelyindependenteventand is does notdepend
1. What do we inheritfromparents?Genotype or Phenotype?
2. Howmanydifferentcombinations of maternalandpaternalchromosomescan be packaged in gametemad by an organismwithadiploidnumber8?

3. WhatwasthereasonforMendelnot to observedeviation in Peaplantexperiment? we findMendelianinheritance in Humanbeingstoo?? Howadominantgeneactuallysuppressestherecessivegene?
Gene is a segment of DNA which carried codes for particular protein
The protein, most of the time is an enzyme which catalyzes the biochemical pathways and result in a phenotype
Thus gene segment is responsible for the phenotype
Gene structure is very simple yet much complicated. Basically it has 3 unite: Cistron, Recon and Muton
Gene fine structure shows the presence of promoter elements which initiates the DNA transcription process. It also has terminator sequences, at the end which help in termination of transcription
The Gene is also said to have other elements: Operator, Repressor, enhancer..etc.
We inherit the genes from the parents and not the phenotypes

Allele
Alleles have similar sequences , but code for variants of enzymes
These enzymes coded by alleles, catalyze different chemical reactions and thus give different phenotypes
Loci

Always alleles of a gene, occupy same loci
Complementation- Complementation is a biochemical phenomenon, in which the gene products from 2 different alleles react with each other to give a complementary product.

The complementary genes cannot produce the phenotype, unless gene product from both the genes is present.
Complementation test is carried out to locate the site of mutation of gene
Pseudo genes- Genes are open for continuous mutation and thus evolution. Throughout the evolution, any time a gene can be mutated, which render it to be non functional. Thus a gene which was functioning in past, may have stopped coding for protein, in recent time. This is called as Pseudo genes.
Processed Pseudo genes- Pseudogenes sometime get converted into mRNA, but fail to form proteins. They get converted to DNA and get inserted into the genome again, as cDNA copies. They do not have any introns.
In prokaryotes the genes is organized into well planned Operons and in eukaryotes similar operons have been discovered recently.
The operons with more powerful sequences wil become the dominant gene and the weak promoters fail to produce their phenotypes and thus get suppressed
It wouldtakeapersontyping 60 wordsperminute, eighthoursaday,around 50 years to typethehuman genome.
We have studied in previous sections (Unit 1), about the structure and function of nucleic acids. They are the building blocks of genetic material, in turn forming chromosomes. We all know that the genetic information is a set of instructions written on the DNA strands. This information is written with only 4 letters, A, T, G and C. They control the type of protein to be formed and the character possessed by the organism. In this chapter we shall try to understand the concept of Gene and its importance. Why always humans give birth to a human baby and a dog always a dog puppy. There is no single example in our long history of life on earth, this pattern in not followed. Why this is so? What is so specific in us and in all organisms that we inherit specific set of characters from our parents? May be there is something in us, which makes us so unique. We call it as gene.
We always say genes make you look this way; genes make you behave that way, but how? What are these genes and how do they make you the way you are? Gene is a very beautiful concept. is just a segment of DNA which has some special codes. Genes code for mRNA and in turn mRNA gets translated into Proteins. These proteins, most of the time are enzymes.
Figure8.6 Gene structure (Basic) Gene is a sequence of DNA which codes for specific proteins. It consists of various sequences elements which help in gene expression, a promoter, UTR, terminator, enhancer…etc


They catalyze some specific biochemical reactions and thus giving a particular final product. That product may be a carbohydrate molecule or a protein moiety or a vitamin complex or a co factor complex or any such chemical molecule. These are responsible for different characters shown by organism Hence we can say, that the genes code for the characters. Gene has very distinct and unique structural features. In brief, it has a promoter which induces the gene expression, it has an enhancer which enhances the gene expression, it has terminators which terminate the genes transcription, and it has ORF (Open Reading Frame) which contains codes for the proteins. This is the basic structure of gene. Gene is also said to have 3 unique components: 1. Cistron (Coding region) 2. Recon (region which undergoes recombination) 3. Muton (Sequence which is open for mutation)
We find in human population that, we do not have same hair color some have black ha have brown hair color and some have red color too. Why so difference, although only one gene cods for this? Le windows case. We have software given by Microsoft, which runs the computer. Compared to a gene We have seen, Windows 2003, Xp professional and then windows vista, windows 7 and now windows 8 also in market. So these all software does the same function running the computer, but different versions, with some different features. The same logic holds for gene and allele Gene is the sequence of DNA coding for the characters.
Figure8.7.Homologouschromosomeshavesimilarstructuralfeatures anddonated by eachparent.Theycontainsame/differentalleles at samelocus. The allele on righthandchromosomecodesforwrinkled seeds in plantsandtheallele on leftsidechromosomecodesforround seeds in plants
Due to mutations a small sequence change happens in sequence and thus the gene now starts coding for altered form of an enzyme. Thus sequence is similar, enzyme coded is also similar, but not the same. Allele is a different form of a same gene.
There is a gene coding for Hair color. It can have many alleles, allele coding for black hair color, allele for brown hair color….etc

As you can see in human population, we have different eye color, black eyes, brown eyes, green eyes, blue eyes…etc. That means there is a gene coding for eye color and it has 4 different allele, each giving different color. Thus the gene can have more than 2 alleles in a population; this is called Multiple allelism. The alleles can be multiple in populations, but any 2 can be in an organism, not all. Because we are diploid in nature, we can only have 2 al
The classical example
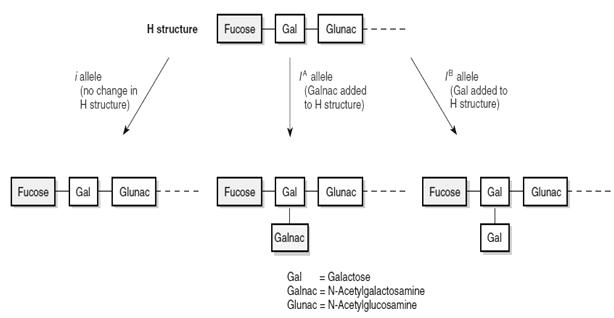
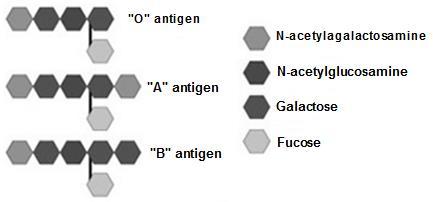
As we all know, we have 4 types of blood groups, 0, A, B, and AB. These are coded by certain alleles, (Refer Figure 8.8)
blood group
blood group
blood group
So, far we learnt that, mutations in alleles/genes give rise to different characters On a same locus, different mutations can happen to give different traits or different mutations can happen at different locus. How do we find it out, what is happening and is the mutation occurred in the same locus or in different gene?
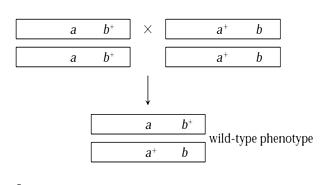
Fruit flies have normal red colored eye. In fruit fly, white is an X-linked mutation that produces white eyes A phenotype Apricot is an X-linked recessive mutation that produces light orange colored eyes. Now, where mutation is occurring? Is it in white locus or in red locus? We shall carry out complementation testing, to figure it out.
Let us, first cross the parents homozygous for different mutations. Figure 8.9, their offspring will be heterozygous If mutations are allelic (occur at the same locus), then the heterozygous offspring have only mutant alleles (ab) and thus become mutants. If mutations occur at different loci, heterozygous offspring will have one mutant and one normal allele at each locus. In this case, mutations complement each other. Heterozygous offspring have wild type phenotype.
Complement group: The genes undergo mutations so that only when they come together, they can give rise to the wild type phenotype. This is what we learnt in Complementation. Let is assume that we have 4 genes, under complementation. Gene 1 can complement with gene2 and gene 2 can complement with gene 1, that means Gene 1 and gene 2 are different. If they were same, they would not have complemented. Thus they both belong to 2 complementation groups.
Consider a hypothetical insect species that has red eyes. Imagine mutations in two different unlinked genes that can, in certain combinations, block the formation of red eye pigment yielding mutants with white eyes. In principle, there are two different possible arrangements for two biochemical steps responsible for the formation of red eye pigment. The two genes might act in series such that a mutation in either gene would block the formation of red pigment. Alternatively, the two genes could act in parallel such that mutations in both genes would be required to block the formation of red pigment. When mutant phenotype is not reverted back to wild type we can conclude that the genes do not complement each other. So, we say they belong to same complement group.
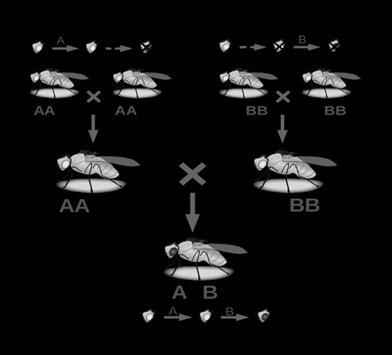
1.Genes belonging to same complementation group cannot complement each other.
2.Genes belonging to 2 different groups can given rise to complementation and final phenotype
The concept of Pseudo alleles or pseudo gene is very interesting. As the name indicates, Pseudo = Fa evolution, the genes have been modified extensively, may be due to mutations, errors during crossing over or errors during replication. Thus these mutations give rise to different alleles. The pseudo allele has sequences that resemble known genes but cannot produce functional proteins. Pseudo genes originate through gene duplication, followed by the subsequent ac-

whenmated gavethewildtype.Whereasthemutants in b) The genes order to give

Figure8.10. The drosophilawithmutantlinescomplement each other to giveahybridred eye color.Presence of both AandB in dominantstate is required to complementthe geneproducts.
cumulation of disabling mutations (e.g., nucleotide insertions, deletions, and/or substitutions) .These mutations can disrupt the reading frame or lead to the insertion of a premature stop codon.
Processed pseudogenes

cDNA copies of the Pseudogene. That means, a pseudogene will be transcribed and it undergoes reverse transcription to form cDNA copy.
again gets incorporated into the genome. This process is called as Retro transposition.
lack introns and a promoter region, but they often contain a polyadenylation signal and are flanked by direct repeats.
in reverse transcription and the lack of an appropriate regulatory environment often lead to the degeneration of processed copies of genes.
appear to have a high number of processed Pseudo gene—approximately 8,000
Drosophila, only 20 retropseudogenes are detectable
most interesting Pseudo gene finding to date is that degenerated protein-coding genes have been proven to "live on" as RNA genes
Pseudo gene can control/regulate parental genes through siRNA
Thus we can say, genomes still remains with all its secrets.
Thus the gene is a mystery yet to solve by us. Everyday new new concepts and phenomenon is getting discovered. There can be more than 10 alleles, giving similar phenotypes and their interaction produces multiple possible phenotypes, this is studied in detail in QTL maps. Complementation helps us to determine how many genes and how many alleles participate in character developing.
is adivisible,yetdistinguishedentity.
whatwaysthecomplementationtestuseful?
is theactualstructure of agene?
In terms of Dominanceandrecessive,whatactuallymeant in molecularterms?
has beensilencedfrommanycenturies, can we call it as Pseudogene?

Mendelian Principles are not valid in every genetic study. We often find deviations from Mendel ratios of monohybrid and Dihybrid crosses. These have given rise to speculations that, the inheritance of genes in not as simple as predicted in Mendel principles.
Mendel gave 2 most important concepts: Law of segregation and Law of independent assortment. The law of segregation is universal law and is applicable in all genetic analysis, whereas the independent assortment law is valid in only special considerations.
Incomplete Dominance – The phenotypes from 2 different genes, blends to give an intermediate phenotype. The ratio of F2 phenotype is 1:2:1
Co-dominance – The phenotype results from two dominant genes existing together .The F2 phenotype ratio is 1:2:1
Gene interactions- The genes in different loci, coding for different phenotypes, interact with each other. One gene acts as a dominant and other acts as recessive, we call the suppression of one gene by the other is called Epistasis.
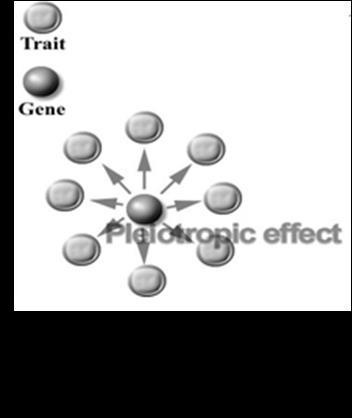
Pleiotropy- One gene, multiple effects. One gene can influence multiple characters, directly and indirectly.
Genomic imprinting – One of the contributing parents influence the genes of the offspring more than the other. It is a type of epigenetic interactions. One of the copies of allele from one parent is made silent and other is always expressed.
Penetrance – The number of offspring, with the genes, expressing the phenotype. Penetrance is said to be 100% , if the genes expresses its phenotype to fullest extent in an organism
Expressivi - The intensity of expression. The gene may be present and expressed in all, but its intensity may vary

Phenocopy – The mimicking of the phenotype produced by environmental causes, of that produced by genotype

Linkage- Genes arranged on same chromosome inherit together to next generation. Thus two genes are said to be linked. Linkage is one of the major reasons for deviation from Mendelism.
Crossing over- The exchange of genetic material between 2 homologous chromosomes, during Prophase –I of Meiosis. Crossing over breaks up the linkage and creates recombination. Crossing over is the necessary drawing force of evolution
Sex linkage
chromosomes inherit these phenotypes. The genes can be on Xchromosome or on Y-chromosomes
Sex linked characters- genes may or may not be present on Sex chromosomes, but the characters are found specifically in one of the sex
Sex influenced characters – The phenotype is found in both the sex, but the intensity or expressivity depends on the sex of the organism
We have been saying and learning so far that the recessive gene is suppressed in presence of dominant gene and only dominant gene character is visible. But sometime, we find the exceptions too. In the Mendelism we saw that, one allele is dominant and the other is recessive. The dominance can sometimes show incomplete domination over the recessive, we call it as incomplete dominance. In the following section we shall analyze some example of incomplete dominance.
Let us take 4’O clock plants with 2 different colors White and red flowered plants, They were crossed to obtain the F1 generation, which were all pink!! Surprise result, the red is dominant over white; still it gave new color Pink. In F2 some were white and half of them were pink flowered. Ratio was 1:2:1 pink; white plant with R1R2 allele has become pink in color. The allele R1 coding for red color is incompletely domi-

havingred whiteflowers.Whenthesetwoplants werecrossed,apinkfloweredplantwasobtained.Redcolorallele
nant over R2 The red and white pigments mixed up to give Pink color. Hence, we can say, in Incomplete dominance mixing of allele products takes place. So, what is happening at molecular and genetic level? The genes Red and White both are expressing and Red gene product is not blocking the white gene product expression, in phenotype. Both the proteins from R and r alleles is mixing up, to give you a new intermediate phenotype.
In human, Tay Sachs disease inheritance follows incomplete dominance.
A defective allele, found most often in populations of Ashkenazi Jews (Eastern Europe), results in a wild type enzyme (hexosaminidase-A) being produced in a non-functional form.
HEXOSAMINIDASE-A is responsible for breaking down lipids. In its absence, sphingolipids accumulate in the developing brain and peripheral nervous system of affected fetuses/young children
Result: brain damage, retardation, death by age five.
TT: normal
Tt: half the amount of enzyme produced, but sufficient for normal development

tt: no functional enzyme; expression of Tay Sachs disease.
This is Incomplete Dominance because both alleles are expressed (i.e., a protein is made from each allele), but only one is functional
This is another deviation from the Mendelism, in which the dominant and recessive genes co-exist and express in full extent together. Thus in heterozygous state, both the alleles will express their gene products and is visible in phenotype. No allele is dominant and no allele is suppressed. Gene products of both alleles get expressed and shown in the phenotype. Ratio of F2 generations: 1:2:1 Classical example is the ABO blood grouping As we learned earlier the ABO blood group is caused by multiple alleles. To form AB blood, both alleles
Figure8.12 The allele ‘A ‘and‘B’bothcoexist in AB bloodtype. Thus boththegeneproductsN-aetylgalactosamineand Galactoseareproduced on the RBC membrane
Unit 8 must be present together. Both allele products are expressed on the RBC. AB blood group causes RBC to have both Galnac and Gal to form on RBC. Figure 8.12 Thus we can see in AB blood group, both the gene products from alleles A and B are present together, without mixing.
So far in our study we have seen that allele of same gene interact. Some alleles behave as dominant over other, or sometimes, both are incompletely dominant and sometimes they show co-dominance. In this section, we shall look into some cases, in which two or more genes are interacting with each other. The interaction between the effects of genes at different loci (not allelic) Gene interaction
Most of the time, one gene masks (hides) the effect of another gene at different locus called as Epistasis. This is similar to dominance and recessive concept, here interacting genes are non allelic and present on different loci. The gene which is masking other is called Epistatic The gene which is getting masked by other Hypostatic gene
Recessive Epistasis (

The concept is very simple. The Epistatic gene (which is dominating on the other) shall be in homozygous recessive state. The Epistatic gene in homozygous recessive state blocks or masks the effect of the other gene. The hypostatic gene (which is
getting masked) can be in homozygous dominant / in recessive state. does not matter, what is the genotype of hypostatic gene. The Epistatic gene should be in homozygous reces-

sive state. Let us look at the below Punnet square in figure 8.13. What can we conclude form this? Who is dominant and who is recessive
Thus gene
Observe the first box
the golden phenotype ‘e’ gene is in recessive condition. And B is in dominant state. What about in Brown state? Brown ‘b’ gene is in recessive condition. And ‘E’ is in dominant condition. Look bbee’, golden color. Here, the ‘ is in recessive condition, and giving golden color. If we say, ‘b’ is Epistatic in recessive condition, then how would you explain the last box. Hence the ‘E’ is in recessive condition, acts as Epistatic.
tion, blocks expression of gene B. Dominant Epistatic. (F2 ratio: 12:3:1)
socialmaturity is verydifficult to measure in men.
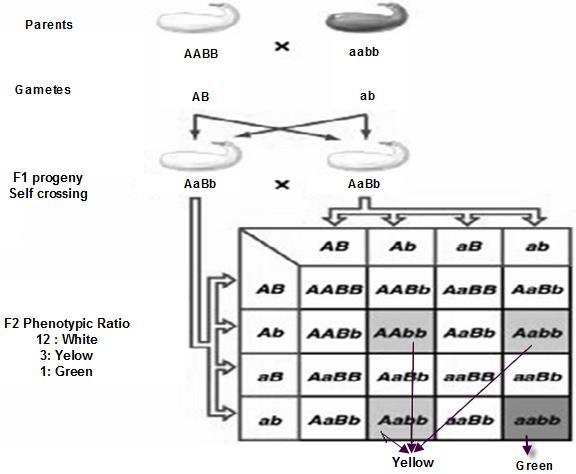
This case is slight variation from recessive Epistasis, as the name indicates the gene in dominant state acts as Epistatic gene The hypostaticgene can be in dominant or recessive state, it does not matter. Let us look at the below chart in figure 8.14 What conclusions can you draw from here?
Whenever, ‘B’ gene is in dominant condition it masks the effect of ‘B’ gene, giving White color.
Whenever, ‘B’ gene has become recessive, ‘A’ gene is not masked and hence we find Yellow color.
When, both the genes are in recessive condition, in last box, it turns out to be green.
Thus B gene is dominantly Epistatic over A gene.
8.3.4 Complementary interactions (Duplicate dominant Epistasis)

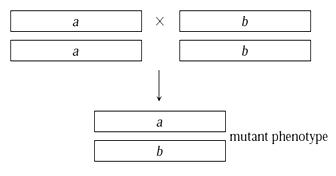
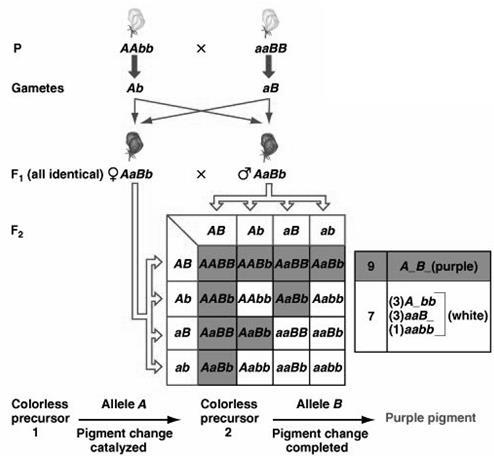
In this type of interactions, both the genes are necessary to show the trait. We have learnt in complementation (Chapter 8.2), that both the allele copies are required to get the result. Similarly, here we need both the genes to get the character. They complement each other. Absence of any one of them shows recessive phenotype. The phenotypic ratio is 9:7, figure 8.15
Genes are present on the chromosomes in an ordered array. Organism has some Autosomal and 1 or 2 allosomes. These contain different sets of genes. Gens present on X chromosome codes for female character, but it also has many other genes which code for normal body functioning. There are certain genes which are very specific for the Sex chromosomes (Allosomes). These traits are passed on from generations through these chromosomes,
either on X or Y. Sex-linked inheritance regularly shows different phenotypic ratios in the two sexes of progeny, as well as different ratios in reciprocal crosses. Inheritance patterns with an unequal representation of phenotypes in males and females can locate the genes concerned to one of the sex chromosomes
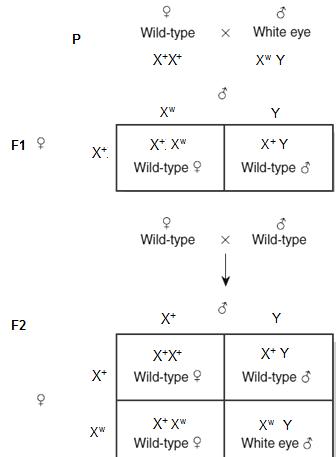
8.3.5.6 X linked inheritance in drosophila

X linked inheritance is caused by the genes present on the X chromosome. X chromosome contains close to 400 genes which code for normal life, other than coding for sexual characters. These were first studied by T.H Morgan in 1910. Let us understand this with the help of the diagram below. (Figure 8.16)
We have crossed 2 drosophila flies, female with Wild type eye (Red color) and male with White eye.
When we self-crossed the F1 generation, interestingly, all females have red eyes. Half of males had red color and half had white eyes.
This is very peculiar because males have 2 different eye colors, and females all have red color.
Figure8. copies abnormalcopy,hencegettingwhiteeye. progeny,themalewithaffectedXchromosome hastheabnormality. The femalewith one affectedXchromosomedoes not showthe disease,because it has one normalX chromosome.
Let’s us study this with the help of X and Y chromosomes
.Thefemaleparenthasbothabnormal Xchromosomesandhence,whiteeyes andMalehasnormalXchromosomeandhence theF2progeny,themale withaffectedXchromosomehastheabnormality. femalewithoneaffectedXchromosome does not showthedisease,because it has one normalXchromosome. The otherfemalewith bothabnormalXchromosomewill be having mutantwhiteeyes. Thus theresult of reciprocal cross is not thesame.
The genes which code for the eye color are present on the X chromosomes. The X chromosomes having Red color allele is dominant over white alleles. (Red alleles X+ and white color (Xw) Females will have 2 X chromosomes and males will have 1 X chromosomes and 1 Y chromosomes. Now analyze the above figure carefully. In the square box we can find, the female have at least 1 X+ chromosome making them Red eyed, in males half have X+ Y making them Red eyed. The other half males have Xw Y making them White eyed. This is how genes present on X chromosomes influence the characters and change the pattern of inheritance.
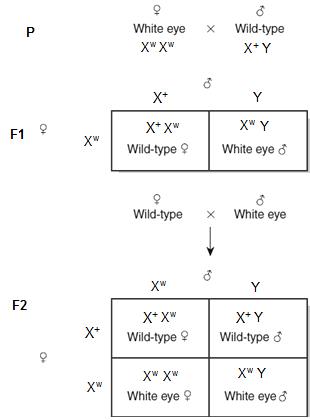
When genes are X linked, the reciprocal crosses show some different ratios, than the normal cross. Let is try to understand this with the help of figure 8.17. Let us continue from our above example of red and white eye drosophila. We had taken female as Red eye and male as white eye. We got all F2 female Red; F2 males 1/2 Red eyed, ½ White eyed Now, lets us exchange the phenotypes of the parents Reciprocal cross: White eyed female and Red eyed male.
In F1 generation: All female Red eyed and all male White eyed. In F2 generation: ½ females White eyes; ½ females Red eye; ½ males White eyes; ½ males Red eye Thus we saw that the reciprocal cross results are not same.

Y chromosome is the male chromosome, found particularly in mammalian males. The genes present on Y chromosome affect on male characters. It is given from father to son and it runs in all males in the family. Only Y chromosome exists in nature and hence there is no chance for crossing over and recombination. The entire length of Y chromosome is heterochromatin and thus it does not code for any proteins. Only a small region on Y chromosome called, The SRY (Testis –determininant region) can code for male characters. Thus maleness itself is Y linked Any mutation in this region can be lethal and passed directly from father to son. Hairy ear case (
Hairs in ears in a very common site in aged people. It is characterized by growth of hair on ears in late adulthood.
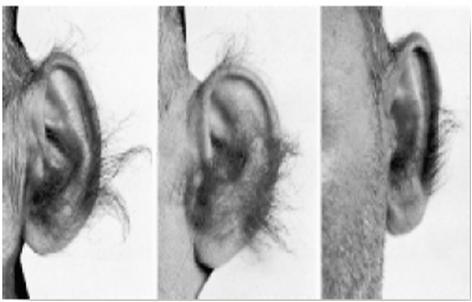
It is seen only in males and never in females. Thus it is thought to be believed due to Y linked gene.
But there has been no direct evidence so far, to confirm that it is Y linked, no gene has been localized to Y chromosome which may cause this condition

It is seen only in Indian population, than any ethnic groups around the world.
Table8.2. Difference between X linked and Y linked and Pseudo
,abelievedYlinkedinheritance, Indianraceandusuallyobserved in aged chromosome human X chromosome and Y
As you know, the males have 1 X chromosomes and 1 Y chromosomes. The X chromosome which is present will express itself and males will get the characters. (Note: X chromosome has several genes which are required for normal body functioning) If the X chromosome gets a mutant allele even that will gets expressed, because males have no other X chromosome to nullify its effect.
Everyhumanspentabouthalf an hour as asinglecell.
Unit 8
Thus we call males as Hemizygous. In the above example of drosophila in figure 8.16 & 8.17: the white eye color is recessive in nature. For a female drosophila to become a white eyed, both of her X chromosomes should have the Xw allele. But males have only one X chromosome makes them more vulnerable. If that gets the white eye allele, male drosophila gets the white eyes. Thus the Xw allele is not dominant in presence of other allele, but behaves as dominant when it is present alone Pseudo-dominance
It is a very interesting case. Our prior genetic knowledge agrees that one gene codes for one enzyme. But sometimes we find that, one gene affecting more than one character at a time! It is called Pleiotropy. A gene is said to be Pleiotropic, if it affects more than one character at a time. Almost all genes are pleiotropic, in nature. All ge
but its protein product, may induce some series of reactions, giving rise to multiple phenotypic effects.( Figure 8.19)
Ex
: 1. Sickle Cell Anemia – Malaria
Sickle cell anemia comes by mutation in the structure of hemoglobin, which makes the structure of RBC to be sickle shaped. This reduces the oxygen carrying capacity of the RBC. But it has very interesting advantage to the person. It makes him resistant for another deadly disease Malaria!
The malaria protozoa survives and reproduces in RBCs. When RBC itself in sickle condition, the protozoan cannot reproduce. Thus the sickle RBC is not favorable place for the protozoan to survive. Thus any person with ‘ss’ recessive condition will be a anemic but resistant to malaria
‘Ss’ heterozygotes are not anemic, but are less prone to malaria

Thus the S allele has direct effect on shape of RBC, but it indirectly affects our resistance to malaria.
2.Marfan Syndrome
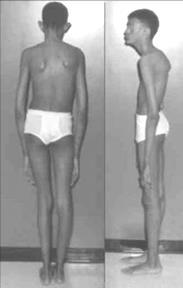
Marfan syndrome is caused by a defect (mutation) in the gene that tells the body how to make fibrillin-1, a protein that is an important part of connective tissue.
The basic features include, long legs and arms, Tall and thin body type Chest sinks in...etc. ( Figure 8.19 A)
This defect results in an increase in a protein called transforming growth factor beta, or TGFβ.
The increase of TGFβ causes problems in tissue throughout the body, which create the different Marfan syndrome features and cause medical problems for people with Marfan syndrome.People can inherit Marfan syndrome, also can be by a spontaneous mutation, people with Marfan syndrome have a 50-50 chance of passing the mutation on each time they have a child.
It does not affect Intelligence
A:The Morphansyndrome showingthin,talllegs andarmsandsticky chest(pectusexcavatum)
It affects multiple body parts[ Phenotypes] including Heart and Blood Vessels (Cardiovascular system), Bones and Joints (Skeletal system), Eyes (Ocular system)

Table 8.3.Prader-Willi Syndrome
Cause Unexpression of 7genesonchromosome15
Symptom
Lowmuscle tone
Short stature
Incompletesexual development
We have seen in our genetic studies, that Autosomal genes are donated by both parents in equal amount. Both participate equally in framing genes of the offspring. But this is not the case, always. For most genes, we inherit two working copies; one from mom and one from dad. But with imprinted genes, we inherit onlyone working copy. Depending on the gene, either the copy from mom or the copy from dad is epigenetically silenced. Silencing usually happens through the addition of methyl groups to DNA during egg or sperm formation. The epigenetic tags on imprinted genes usually stay put for the life of the organism. But they are reset during egg and sperm formation. Regardless of whether theycame
Thus only one copy of thegenes and thus reduced number of transcripts
from mom or dad, certain genes are always silenced in the egg, and others are always silenced in the sperm
We find that one of the parent influences the genotype of the offspring more than the other parent. This differential expression of genetic material depending on whether it is inherited form male or female parent
The imprinting is achieved by 2 basic mechanisms.
1.Covalent
Thus only one copy of the genes and thus reduced number of
(DNA: DNA, DNA: Protein interactions). In mice, p ternal imprinting favors the production of larger offspring, and maternal imprinting favors smaller offspring. Thus the paternal gene try to absorb more nutrient form mother during development and mother genes tries to counteract it. Maternal genes try to reduce maternal investment on the child. This result due to mother tries to limit the size of the mice, because to make the mice size larger, it needs to give more nutrient. Father tries to make the size of offspring larger, because he does not give nutrition to growing embryo, no loss for him. You're not an equal product of both parents' genes.
The phenomenon of genomic imprinting evolved in a common ancestor to marsupials and Eutherian mammals over 150 million years ago. Thus, genomic imprinting evolved in mammals with the advent of live birth. Its evolution apparently occurred because of a parental battle between the sexes to control the maternal expenditure of resources to the offspring.

Genes are the manifestations of the environmental causes. Organisms are in constant interaction with surrounding environment. The environmental stress and requirements shape the organism and expression pattern of the genes. These interactions affect the behavior of the organism and thus its survival. Environment includes temperature, rainfall, extreme weather, nutrition, availability of food and water, presence of predators, competition…etc.
The phenotypes molded by environment resemble that of the phenotype coded by genes. Thus the genes are not absolutely required to produce the phenotypes. This is the characteristic of Phenocopy. It creates problems during pedigree analysis and other genetic disorder studies. A human being whose hair has become lighter in shade due to prolonged exposure to bright sunlight, as opposed to another whose hair is naturally that color. An animal whose coat changes color with changes in temperature is another example. Snowshoe hares have a coat that becomes white during the winter but reverts to mottled brown and grey shades during the summer. I Rabbits, Himalayan allele produces dark fur near nose ears and feet.
temperaturesensitive.Rabbits patches at higher temperatureandnot
When the rabbits are reared at 25°C or less, they develop dark pigments
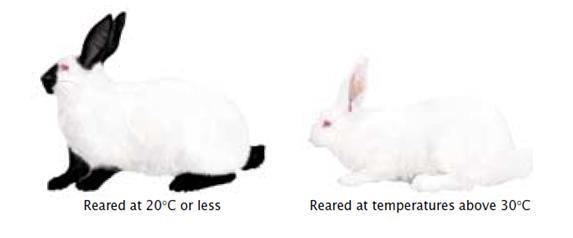
When rabbits are reared at 30°
Thus the expression of Himalayan allele is temperature dependent, also called as Temperature sensitive allele
Genes and their proteins do not act in isolation.
They are in constant influence form the environment
Sometime, environment can alone produce a phenotype, exactly as produced by a gene called Phenocopy
are solely induced by environment
are
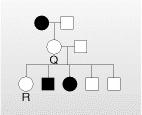
We have assumed so far, that any organism with the given genotype, expresses the phenotype. That means if 100 people have a genotypes, then all should show the phenotype. But, it is not always true. Sometime an organism may have the genes but fail to express them, even though genes are in dominant state. Thus the gene has to penetrate in the organism to show its phenotype. This is called as Penetrance. Penetrance is the percentage of individuals having a particular genotype who express the associated phenotype. For ex: If we have 100 people with given gene A, 90 of them will show its phenotype. That means Penetrance is 90% Reduced Penetrance: If some people with the mutation do not develop features of the disorder, the condition is said to have reduced (or incomplete) penetrance. Reduced penetrance often occurs with familial cancer syndromes. For example, many people with a mutation in the BRCA1 or BRCA2 gene will develop cancer during their lifetime, but some people
Figure8.21.Lack of penetrance illustrated by apedigreefora dominantallele.IndividualQ musthavetheallele(because it waspassed on to herprogeny), but it wasnotexpressed in her phenotype. An individualsuch as Rcannot be surethather genotypelackstheallele.
will not. Doctors cannot predict which people with these mutations will develop cancer or when the tumors will develop. Reduced Penetrance probably results from a combination
of genetic, environmental, and lifestyle factors, many of which are unknown. This phenomenon can make it challenging for genetics professionals to interpret a person’s family medical history and predict the risk of passing a genetic condition to future generations. Figure 8.21 If all organisms do not show the expected phenotype, we call it as Incomplete Penetrance. In general, when we know that the genotype is present but the phenotype is not observable, the trait shows incomplete Penetrance. Basically, anything that shows less than 100% Penetrance is an example of incomplete Penetrance. A specific example of incomplete Penetrance is the human bone disease Osteogenesis imperfecta (OI), figure 8.22. The majority of people with this disease have a dominant mutation in one of the two genes that produce type 1 collagen, COL1A1 or COL1A2. Collagen is a tissue that strengthens bones and muscles and multiple body tissues.
Symptoms:
Weak deformed
Bluish color in the whites of their eye
Variety of afflictions that cause weakness in their joints and teeth.
However, this disease doesn't affect everyone who has COLIA1 and COLIA2 mutations in the same way. In fact, some people can carry the mutation but have no symptoms. Thus, families can unknowingly transmit the mutation from one generation to the next through someone who carries the mutation but does not express the OI phenotype.

Although we all have black hair color, but the intensity of the color is different. Some have dark black and some have light black color. This means the intensity of the expression is different. This we call as Expressivity Expressivity is the degree to which a trait is expressed. Individuals with the same genotype can also show different degrees of the same phenotype. Expressivity is the degree to which trait expression differs among individuals. Unlike Penetrance, expressivity describes individual variability, intensity of expression.
Although some genetic disorders exhibit little variation, most have signs and symptoms that differ among affected individuals. Variable expressivity refers to the range of signs and symptoms that can occur in different people with the same genetic condition. For example, the features of Marfan syndrome symptoms (such as being tal others also experience life and blood vessels. Although the features are highly variable, most people with this disorder have a mutation in the same gene FBN1 Another example of expressivity at work is the occurrence of extra toes, or polydactyl, in cats. The presence of extra toes on a cat's paw is a phenotype that emerges in groups of cats who have interbred for generations. In fact, there are several well-known groups of these cats, such as those on Key West Island (known as "Hemingway's cats"), as well as those in breeding clusters in the eastern U.S. and shores of the British.
The first to report on this phenomenon was C. H. Danforth, who studied the inheritance of polydactyl among 55 generations of cats. He observed that the polydactyl phenotype showed "good penetrance, but
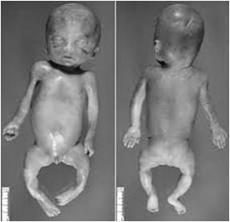
Figure8.23.Variable expressivityshown by 10 grades of piebaldspotting in beagles
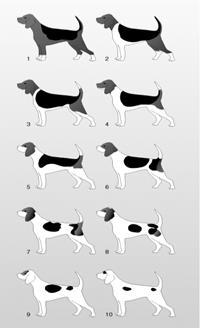
variable expression" because the gene always causes extra toes on the paw, but the number of extra toes varies widely from cat to cat. Through his breeding studies, Danforth found that although a dominant allele underlies the cause of polydactyl, the degree of polydactyl depends on the condition of adjacent layered tissues in the developing limb; that is, the expression of genes in tissues surrounding tissue that will become the toe determines the degree of polydactyl As you can see in figure 8.23, each of these dogs has SP, the allele responsible for piebald spots in dogs. As with reduced Penetrance, variable expressivity is probably caused by a combination of genetic, environmental, and lifestyle factors, most of which have not been identified. If a genetic condition has highly variable signs and symptoms, it may be challenging to diagnose.
Genetics is the most fascinating subject, man has ever discovered. It is a story, a very long story indeed Story which started by the first organism on earth and it continues till today. Characters, features and lot more is getting transferred from one organism to the new individuals. This process is working perfectly since millions of years and will continue for next billions of years till the end of life on earth.
samechromosomearelike2 rope.Wheneverthefirstballgoes,other
Let us see how these genes (which bring characters to you) are located on chromosomes….Genes are just like 2 boys who are sitting on a chair. Problem is that they are tied together by rope very tightly Now they cannot move away, without breaking the rope. Even if you pull them apart, they will move together. We say, these 2 boys are linked or joined together.
This is the concept of linkage mosome tend to move together to next generation. Thus the genes located closer have higher chance to be inherited together. Linked genes tend to stay together and thus increase the parental types.
The genes present on same chromosome are usually linked to one another…but, crossing over (which involves exchange of genes/alleles) breaks the linkage.
As you can see in the figure 8.24 A, the F1 ‘AaBb’ organism produces 4 types of gametes: in that 2 are (AB and ab) are also produced by the parent plants. Hence we call them as parental gametes. The rest 2 ( Ab and aB) are not produced by parents and hence they are new combinations, arising due to crossing over. They are called as Recombinants.
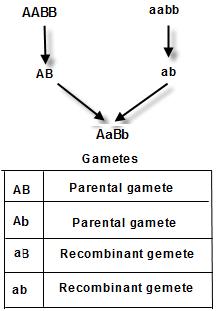
Table8.4.whengenesarenotlinked, andrecombinantgametes.
Parental
Kind of parents
Ex: Ab aB
Parenta type = 50%
Ex: ab AB
Recombinant type
Recombination is the sorting of alleles into new combinations.

Interchromosomal recombination, produced by independent assortment, is the sorting of alleles on different chromosomes into new combinations.
The cross betweenthe‘AABB’and‘aabb’ producedprogenywith‘AaBb’ genotype.Out of the4 gametesproduced by it,2are exactlysame as produced by theParents.Hencetheyare called as parentaland2are newcombinations,called as Recombinants
Intrachromosomal recombination, produced by crossing over, is the sorting of alleles on the same chromosome into new combinations.
In any cross, if number of parental gamete is more than the recombinant ones, then definitely it is linkage.
Imagine the genes A and D are separated by 2 genes, Abcd// ABCD. Gametes Produced by crossing over (Exchanging one gene at a time) ; ‘AbcD’ and ‘ABCD’ with no cross over ;’ AbcD’ and ‘AbCD’ by cross over between B//b ; Again we can get 2 more by crossing over involving both ‘B’ and ‘C’ genes. Thus we may get 4-6 gametes.
Now, Imagine gene ‘A’ and ‘D’ are separated by 4 different genes: ‘Abcefd’ // ‘ABCEFD’ Gametes produced by crossing over, exchanging 1 gene at a time. AbcefD and ‘ABCEFD’ with no cross over ‘ABcefd’ and ‘AbCEFD’ by cross over between B//b; ‘AbCefd’ and ‘ABcEFD’ by cross over between C//c; thus we can have many more gametes by cross involving all sets of genes between ‘A’ and ‘D’
Hence we can assume that, as the distance between 2 genes increases, the crossing over will be more and hence more will be recombinants. (Refer Figure 8.25)
Greater the distance between linked genes,
Greater the chance of cross over, Greater will be number of recombinants.
Unit of gene distance is cM ( Centi Morgan) or mu ( Map Units)
Highest possible frequency = 50% (In case of Mendelism), that means the genes are either on 2 extreme ends of chromosome or on different chromosomes.
If a cross produces 2 recombinants in 10 progeny, for 2 genes A and B. How far the 2 gene are located?
Solution:
Its frequency of recombination = 20%. What That means the 2 genes are 20 m.uapart
The cross-over percentage between linked genes A and B is 40%, between B and C is 20%, between C and D is 10%, between A and C is 20%, between B and D is 10%. What is the sequence of genes on the chromosome?

Solution:
A and B are 40% 40 m.uapart.
C and D 10% 10 m.uapart
B and C are 20% 20m.u apart
A and C 20% 20 m.uapart
B and D 10% 10 m.uapart.
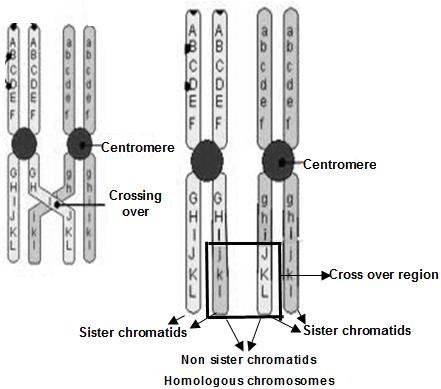
The genes on same chromosome are linked and tend to be inherited together. That means all the progeny should be of same type and shall resemble the parents always. But this is not the case in nature; we have all possible combinations and phenotypes. How this is achieved? The molecular event which brings about the changes in gene combinations and breaks up the linkage is called as Crossing over. Crossing over is an event occurring during Pachytene stage of Prophase-I of meiosis, which begins with the chiasma formation and exchange of genes along homologous chromosomes. The genes on non get exchanged
Molecular details are dealt in Unit 3, Chapter A Recombination
Crossing over is a hypothesis, which is believed to occur during the chiasma formations. We cannot see the movement of genes and alleles but we only can find out the final outcome. The chromosomal and DNA sequences responsible for activation and control of cross over are still a mystery. There are many protein complexes which play role n crossing over.
1. SC proteins
It is a protenaceous complex, which binds to entire length of chromosomes types of proteins with varied functions. Major building blocks of complex are proteins
plex) promotes ZIP1 polymerization

There are 4 major ZIP proteins: Zip1, Zip2 Zip3, and Zip4. Mutation in Zip2 and Zip4 results in complete lack of SC formation. Zip 1 protein, promotes centromere coupling and promotes initiation of synapse. ZIP 1 Represses crossing over near centromere. Thus centromere remains constant entity in chromosome and is not open for recombination.
Crossing over is the main phenomenon behind the linkage. Linked genes do not allow the Chiasma to form between them, and thus tend to increase the parental type progeny. The crossing over is a theory which explains the recombination event, but there is no strong proof of the molecular events which bring about the process.
‘
homologous allelespresent on the thedriving
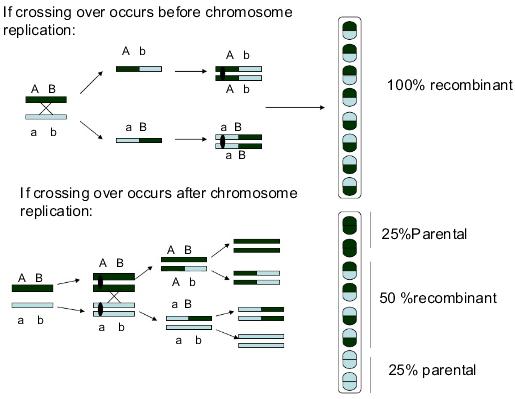
Figure8.27. In case of neurospora, Crossovercanoccurbefore DNA replication or afterchromosomalreplication. If thecrossoveroccurs beforethechromosomereplicates,alltheprogenywill be recombinant ones. Ifit occursafterthechromosomalreplication,then 50%will be recombinants

If geneAandgeneBarelinked obtainingthehomozygousgametesforthegeneAand
How

are 2 types of mapping techniques: 1. Genetic linkage maps 2. Physical maps
maps are the relative distance of genes on the chromosomes
the linkage maps, the distance is measured in terms of recombination frequency.
is not real time distance , but only relative distance between 2 or more genes
possible distance on chromosome is 50 cM.
and DCO are the products of cross over
DCO occurs with very low frequency
SCO should occur together to give one DCO

SCO blocks occurrence of DCO
under suppression, some DCOs still occur

Coefficient of Coincidence
Tetrad analysis is done in fungus, Neurospora and yeast
Neurospora has 2 rounds of meiosis and mitosis
Ordered tetrad
between gene and centromere = cross over / total x 100
distance = ½ ( the above frequency)
Unordered tetrad
PD, NPD and TT
Somatic cell hybridization helps to locate the gene on chromosome.
Molecular markers enable us to identify the gene location and its linkage patterns.

Genes are the discrete unites of DNA sequences, spread all over the chromosomes. The location of gene on a chromosome is very specific. Genes do not change their locations, unless moved by transposition. Genes can be mapped to chromosomes and exact part on the chromosomes using several techniques. In this chapter we shall understand the molecular details about various mapping techniques.
In the previous section we have seen the concepts of linkage and how to determine the distance between 2 genes. In this section we shall try to analyze more complicated gene interactions and mapping techniques. construct a genetic map, by a series of testcross for pairs of
genes, We use a very efficient method called three-point cross, to analyze more no. of genes and to construct maps.3 point means, we establish order of 3 genes. Let us consider a crossing over event, as shown in the diagram below
The parent chromosome is a heterozygote : ‘AaBbCc’
The top of the parental cell, with tetrad stage.
Genes are in coupling configuration on one chromosome and ‘abc chromosome

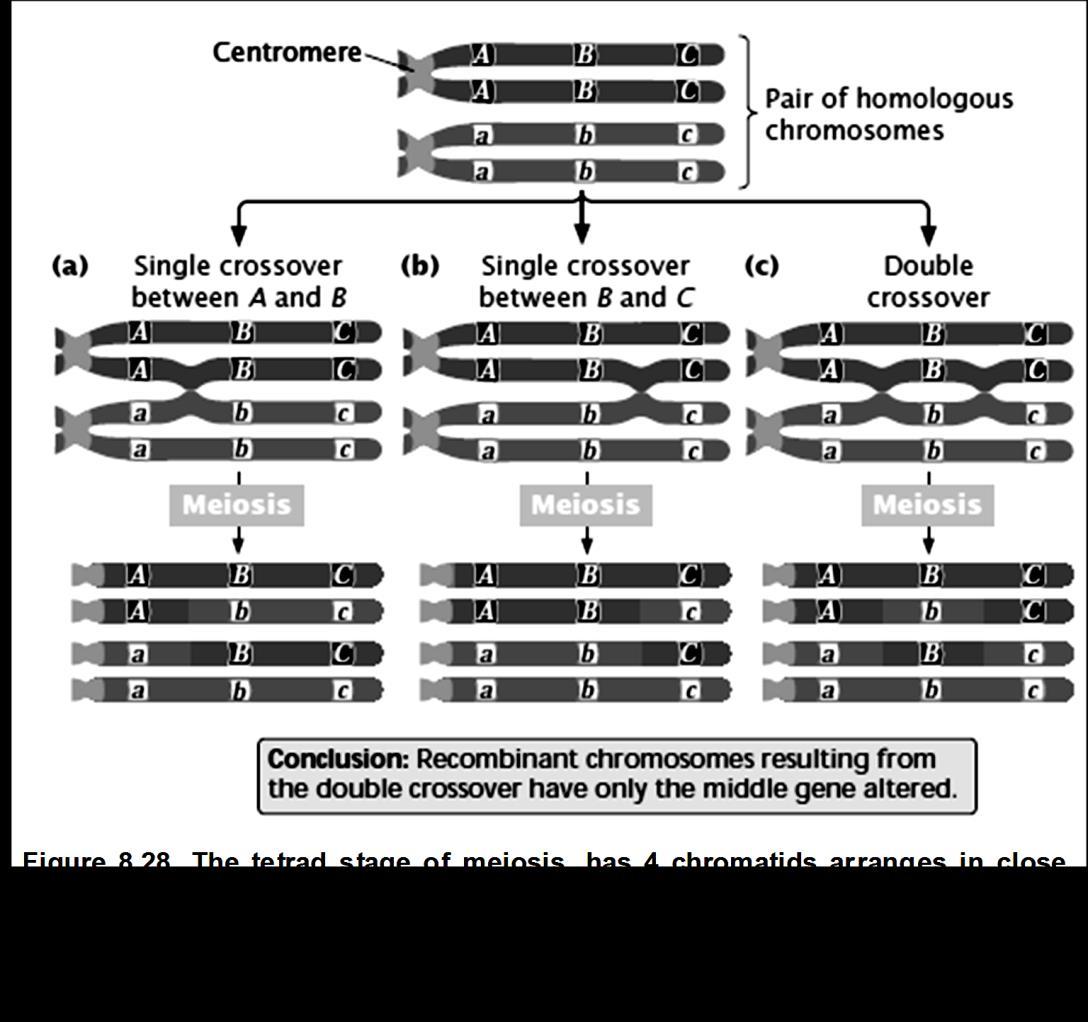
When all dominant ( wild type alleles) are on one chromosomes and all the mutants (recessive) on the other, we call it as Coupling
We can have 2 single cross over and 1 double cross over.
In diagram (a) 2 gene is getting exchanged BC/bc
In diagram (b) 2 genes are getting exchanged BC/bc but the location of crossing over event is different.
In diagram (c) only the middle gene is getting exchanged.
If 3 genes are given ABC, we need to find out the middle gene to construct the map. I. To determine the middle locus in a three-point cross, compare the double-crossover progeny with the non-recombinant progeny.
II.The double crossovers will be less in number.
III.The non-recombinant (Parental) will be the two most-common classes of phenotypes.
IV.The double-crossover progeny should have the same alleles as the nonrecombinant types at two loci and different alleles at the locus in the middle. Let us study this with the help of a sample problem. In D. melanogaster, cherub wings (ch), black body (b), and cinnabar eyes (cn) result from recessive alleles that are all located on chromosome 2. A homozygous wild-type fly was mated with a cherub, black, and cinnabar fly, and the resulting F1 females were testcrossed with cherub, black, and cinnabar males. The following progeny were produced from the testcross: Determine the order of the genes on its chromosome.
Solution:
Determine the parental types
Parental types are found in maximum frequency.
Double cross over recombinants
Double cross over will be the lowest 2 frequencies.
Compare the double cross progeny with parental types.
All 2 alleles will be same, except one allele.
That allele should be in middle.
Parental types:
Double cross over: Compare these 2
1 cross progeny change. That means ‘cn’ is in the middle.
Because in DCO, only the middle gene will be exchanged ch+ cn+
Now how to find the distance between each gene
the single cross over products.
Single cross over between first 2 genes and check the progeny obtained in the table. Count the recombinant number and thereby find the frequency
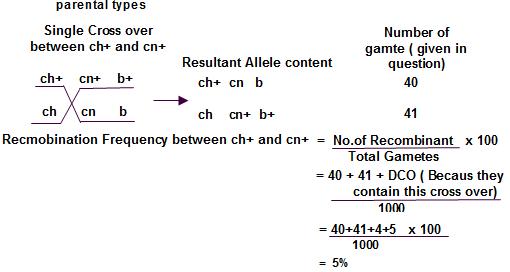
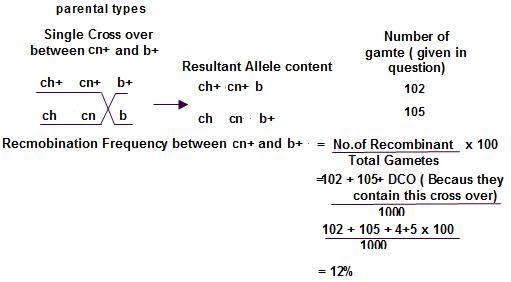
Similarly single cross over between the next 2 genes and check the progeny obtained. Calculate the recombination frequency.
count DCO while calculating the Recombination frequency.

chromosomes
recombination frequency = the distance on the genetic map. 1% frequency means the distance between the genes is 1 cM .So, we got answer as 7%, means distance between ‘ch’ and ‘cn’ is 7cM
Unit 8
Similarly, in second case, the cross over occurs between, the ‘cn’ and ‘b’ genes. The frequency is 12% and hence the distance is 12cM Interference
We have learnt so far about SCO and DCO formations Probability rule says, the probability of DCO = Probability of both the SCOs occurring together! Because, in DCO, we find both the SCOs occurring together, right? Hence their probability should be same. Let us consider an example: DCO = 10; SCO = 280 SCO between h+ and th+ = 200 (you know how to find it out) (add DCOs also)
Probability = 200/1000 = 0.2 SCO between th+ and ry+ = 100

Probability = 100/1000 = 0.1
Probability of DCO = Probability of SCOs

So, we had to get 20 DCOs. How much we getting
That means, something is not allowing the DCO to occur.
We can say, the First SCO, is not allowing the second SCO happen along with that.
When both occur together we get DCO, right!!
Thus the first crossing over is suppressing the other ference
The extent of interference = no. of observed DCO no. of expected DCO
Inference = 1 - Coefficient of Coincidence
Gene mapping is the techniques, which has no standard or single method in every organism. In this section we shall study, some important aspects of mapping in Fungi :Neurospora and Yeast, Neurospora
shows 2 rounds of meiotic events followed by a single mitosis. Thus it produces 8 cells at the end of reproductive cycle. Before mapping any gene on chromosomes we need a reference point, from which distances are measured. In any maps, centromere serves as a good reference point, as centromere does not show crossing over and shift in position. Thus we take a gene of interest and centromere and check the distance between them. Once the gene distance has been establishes,

Observed is less Observed is more
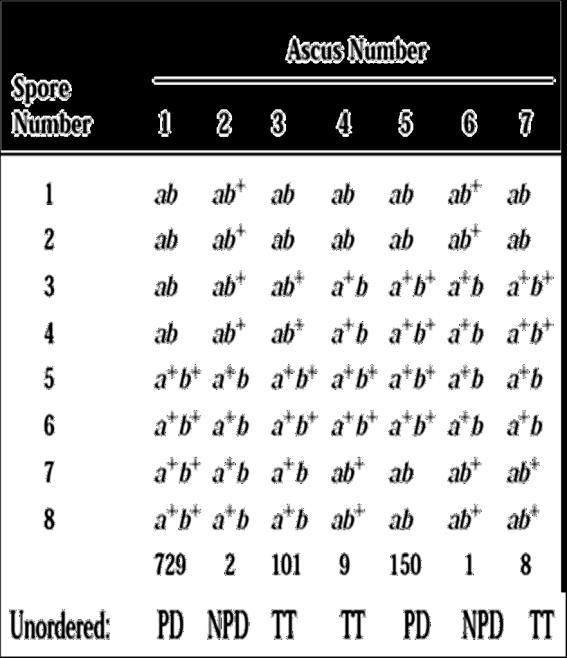
then we can measure other genes distance from the gene of interest, very easily. Depending on this, there are 2 basic types of mapping: Ordered and Unordered maps.
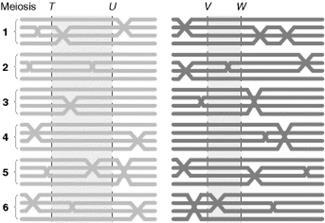
8.4.2.1 Ordered tetrad.
If we know, a gene, say ‘A’ on chromosome, we need to find out its location on that chromosome We shall consider Centromere as reference point, hence we say, the gene ‘A’ is this much distance away from centromere.
Unit 8
As you can see below figure 8.30, the parental combinations come when crossing over does not take place. Thus the “A” and “a” alleles separate during Meiosis I I division segregation. The cross over products, arise when crossing over exchanges the chromatids. Here we, find that the alleles “A” and “a” segregate during Meiosis II, figure 8.31, the 4 type of spores are formed depending on the cross over types and chromosomal arrangement during metaphase of meiosis. The ratio has changed and it is now 2:4:2 and 2:2:2:2
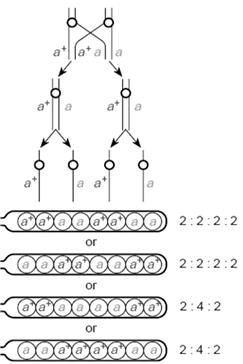
Now, how to find out the distance between centromere and the gene “A”?
Formula: over products / Total no. of offspring x 100
Figure.8.32 groupsareformed.PD,NPDand no crossover,NPDresultswhenallchromatids undergocrossoverandTetratyperesultswhen2 the4chromatidscrossover
Thus distance between the gene and centromere is 7 mU
Always, remember to half the value of frequency, in dealing with ordered tetrad. Ordered Tetrad is shown by Neurospora.

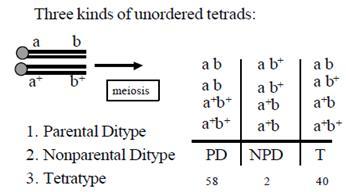
This type is shown by Yeast. It does not produce 8 cells after the reproduction, rather stops at meiotic 4 haploid cells. Let us see, as in figure 8.32, when 2 genes are there, and we need to map the distance between them. This is called as unordered tetra analysis. The parental types are ‘ab and ‘a+b+’ combination. Nowlook at the column written as PD; first spore is ‘ab’, parental type, next is also ‘ab’; then we see ‘a+b+’, parental and last one also parental. Thus it has only parental type gametes and hence shows no cross over. We call it as Parental Ditype
division segregationwithcross overbetweenalocusand centromere
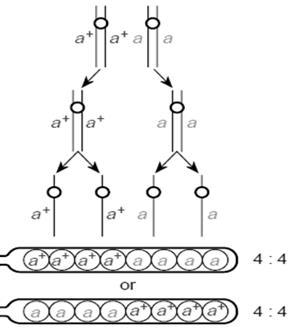
Weknow that when cross over occurs; only 4 chromatids participate and only 50% will be recombicombinants.
FirstDivisionSegregation: Crossingoverdoesnotoccur. parentaltype of
Figure8.30.Thealleles‘a+’and‘a’are on homologouschromsomes. Whenthey do notcrossover,thesegregationoccurs in meiosisI. The raio of gametes at theend of reproductivecycel is 4:4. No recombinantsareborn intheprocess
Unit 8
Observe the next column ‘a+b’ and ‘ab+’, both are recombinant and non-parental ones, Hence the name Non parental Ditype
Last column has 50% parental gametes and 50% recombinant gametes. Tetra type (TT)
Thus when 2 genes are involved we get PD, NPD and TT
Distance between the gene A and B = ½ (40) + 2 x 100 / 100 = 22 mU : Very simple right!!!!!!!!
Let us look at some sample problem
Find the distance between the genes a and b, in the adjacent table:
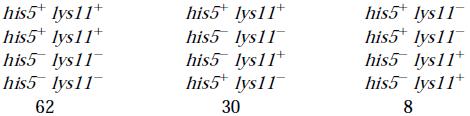

Solution
Find out is it Ordered / Unordered Determine PD, NPD and TT ( parents types: ‘a+b+’ and ‘ab’)
The ad5 locus in Neurospora is a gene for an enzyme in the synthesispathway for theDNA base adenine. A wild-type strain ( ad5. The diploid undergoes meiosis, andonehundred asciare scored for theirsegregation patterns with thefollowing results: What isthedistance of thead5genefrom centromere?
You know the rule, refer the above explanation.
Calculate the total tetratype: 101+9+8 = 118
Calculate the total NPD: 2+1 = 3
Distance = (1/2)Tetratype+NPD/ Total gamete
Solution:
As we can clearlysee, first 2 rows are parental ones andrest 4 iscross over products. Hence distance = cross over productsx 100/total asci = 14% = half of it is recombinant Hence distance between gene and centromere = 7%
In yeast, the in the synthesis pathway for the amino acid histidine, and the synthesis pathway for the amino acid lysine. A haploid wild-type strain (his5 lys11) is crossed with the double mutant ( meiosis, and 100 asciare scored with the following results.
Solution:
PD column 62 ( all are
NPD column 8 ( all are his5+ lys11-and his5-lys11+ gametes) TT column 30 ( mixture of parent and recombinant)
Distance = 1/2 (30) + 8 x 100 / 100 = 23 mU
We cannot locate all genes on the chromosomes. We need help of some other factor to locate each gene. It is similar to a land mark, when we are looking for some address on the map. In this section, we are going to analyze the land marks that we use to map the genes on chromosomes. Genetic land marks are called Markers. Markers are usually some genes or repeat sequences. We use 2 basic types of molecular markers based on nucleotide differences and those on differences in the amount of repetitive DNA.

Unit 8
Some positions in DNA are copied by a different nucleotide in different homologous chromosomes. These differences are Single nucleotide polymorphisms (SNPs) Mostly they are silent, and provide markers for mapping We can detect the presence of molecular markers using restriction analysis. We use these locations to map the gene of our interest.

In many organisms some DNA segments are repeated in tandem at precise locations. But, the precise number of repeated units in such a tandem array may be variable Variable number of tandem repeats (VNTR) we use a probe that will bind to the repetitive DNA and thus helps in location. The other three most common types of markers used today are RFLP, RAPD and isozymes.
RFLP - Restriction Fragment Length Polymorphism; a molecular marker based on the differential hybridization of cloned DNA to DNA fragments in a sample of restriction enzyme digested DNAs; themarker is specific to a single clone/restriction enzyme combination
RAPD - Randomly Amplified Polymorphic DNA; a molecular marker based on the differential PCR amplification of a sample of DNAs from short oligonucleotide sequences
Isozyme - A molecular marker system based on the staining of proteins with identical function, but different electrophoretic motilities
Segregating population is analyzed by RFLP, RAPD or isozyme makers. All of the segregation data is then compiled and used to derive the linkage relationship among the markers. This analysis is performed using computers and one program widely used is called MAPMAKER. This procedure is based on the maximum likelihood method. The output from this program is a linear relationship among the markers and the distance between the markers is measured in centimorgans.
In the first 70 years of building genetic maps, the markers alleles producing detectably different more and more researched, large numbers of such genes could be used as markers on the maps. However, even in those in which the maps appeared to be “full” of loci of known phenotypic effect, measurements showed that the chromosomal intervals between genes had to contain vast amounts of linkage analysis, because there were no markers in those regions.What was needed were large numbers of additional genetic markers resolution map. This need was met by the discovery of various kinds of molecular markers. What can be the reason for the changes at allel
1.Polyploidy (changes in number of chromosomes)
2.Gene or point mutations
3.Recombination
4.Changes in chromosome structure
5.Transposition: mobile genetic elements

A molecular not associated with any measurable phenotypic variation.
Such a “DNA locus,” when heterozygous, can be used in mapping analysis just as a conventional heterozygous allele pair can be used. Molecular markers can be easily detected and are numerous in a genome. Note that, in mapping, the biological significance of the DNA marker is not important in itself; the heterozygous site is merely a convenient reference point that will be useful in finding one’s way around the chromosomes. In this way, markers are being used just as milestones were used by travelers in previous centuries. Travelers were not interested in the milestones (markers) themselves, but they would have been disoriented without them.
Figure8.33 A: On aSouthernhybridization of an individual, heterozygousforthemarkers,theprobewouldhighlightthree fragments, of size3,2,and1 kb
individual, homozygousforthemarkers,theprobewouldhighlightonly1
The two basic types of molecular markers are those based on restriction site variation and on repetitive DNA Molecular or DNA marker techniques allow gene mappers to directly analyze DNA and observe allele differences as specific loci. A genetic marker is any gene that has been mapped. In our examples so far, alleles of the genes we map control phenotype differences we can see expressed in the organism. Molecular markers or DNA markers are variable DNA sequences that have been mapped to a position on the chromosome. The analysis of DNA marker data to determine linkage maps works the same as with genes that control phenotype differences. The advantage of DNA markers is that geneticists can find many DNA differences between parents and track the inheritance of hundreds of marker loci in a single cross. Later we will learn about the details of the DNA analysis techniques that are used to generate a marker phenotype or DNA fingerprint. For now we will go through an example of how geneticists work with this
Use of restriction fragm
Bacterial restriction enzymes cut at specific target sequences that exist by chance in the DNA of other organisms. Generally, the target sites are found in the same position in the DNA of different individuals in a the DNA of somes. However, quite commonly, a specific site might be missing as a result of some mutation. The mutation might be within a gene intergenic area heterozygous for presence and absence (+/−), that locus can be used in mapping. The derived from DNA of that region.
C.Crossoversbetweenthesesiteswould d–2–1. In or to other
The figure 8.33 A and B, clearly indicate the effect of restriction sites on the probe and bands obtained. These multiple forms of this region constitute a In a cross of the afore
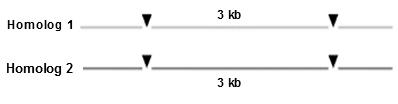
he progeny would show three fragments when probed and the other half only one fragment, following Mendel’s law of equal would. [Note: Try to connect with the mating of heterozygous individuals, AA and Aa, we can have AA and Aa progeny This is what we meant, by heterozygosity in molecular level, isn’t it fascinating?] RFLP can be mapped and treated just like any other chromosomal site. The following situation shows of the heterozygous RFLP in our example to a heterozygous gene, with D in coupling conformation with the 1 plus 2 morph: Figure 8.33 C


The number of repeated units in a tandem array is variable. The important fact is that individuals that are heterozygous for different numbers of tandem repeats can be detected, and the heterozygous site can be used as a marker in mapping. A probe that binds to the repetitive DNA is needed. The adjacent example uses restriction enzyme target sites that are outside the repetitive array. The basic unit of the array is shown as an arrow.
Electrophoresis Geneticists use a DNA separation technique called electrophoresis to detect DNA differences at a particular locus. An individual’s DNA is loaded into a lane in the electrophoresis system and the pattern of DNA bands in that lane is their marker phenotype.
If the DNA sequence at a particular chromosomal region is identical among all members being tested, only one banding pattern will be observed. In this case, the marker phenotype is called Monomorphic.

If the DNA varies, the marker banding pattern will vary across the lanes of DNA from different individuals and we have a Polymorphic marker locus.
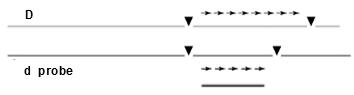
Each marker phenotype can be designated as an allele at that locus.
We have an example of one parent that is homozygous for the 'a' allele and the other parent homozygous for the 'A' allele. We see only one band in the parent lanes because both of their chromosomes have the same type of DNA at that locus.
However, the F1 offspring has a two banded pattern.
The F1 inherited the 'A' allele from one parent and the 'a' allele from the other. Thus these DNA markers can have a lack of dominance and heterozygous individuals can be distinguished from either homozygous genotype. The remaining lanes show the marker phenotype of several test cross progeny produced by crossing Aa with aa. All the progeny have the 'a' marker from the “ but some inherited the 'A' and others the 'a' from the 'Aa' F1 parent. For example, the marker genotype for the first 3 testcross individuals would be aa, Aa, aa. If a DNA marker locus is linked to a gene controlling an important trait, the marker and the trait will tend to be inherited together.
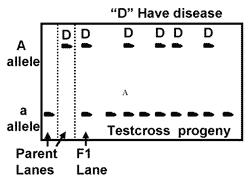
In experimental is facilitated not only by the availability of readily visible markers but by the ability to carry out specifically desired genetic crosses and to obtain large numbers of progeny from any given cross. In human genetic studies, few visible markers are available and planned crosses are not possible, so a different approach is required to map mutations relative to known markers.
In this approach, families in which individuals are at risk for a genetic disease (i.e., both parents are heterozygous for Autosomal DNA samples from various family members are analyzed to determine the frequency with which specific polymorphic markers segregate with the mutant causing the disease; this frequency is a measure of the distance between the markers and the mutation The more families afflicted with a particular disease that are available for study and the more DNA polymorphisms identified in proximity of the disease locus, the more precisely the disease gene can be mapped.
In most family studies, however, locating more than 100 individuals is highly unlikely, limiting the localization to no less than 1 centimorgan. An alternative strategy, which can be used in some cases, is called linkage disequilibrium. In this approach one assumes that a genetic disease commonly found in a particular community results from a single mutation many generations in the past. This ancestral chromosome will carry closely linked markers that will be conserved through many generations.
Markers that are farthest away on the chromosome will tend to become separated from the disease gene by recombination, whereas those closest to the disease gene will remain associated with it.
E.Electrophoresis gelshowingtheDNAmarker parentsandFigure8.33 D:The no. of repeatsmayvary in alleles, ThisVNTR[VariableNumber of Tandem Repeats]locuswillformtwobands, one longand one short, on aSouthern hybridizationautoradiogram.Onceagain,this mappingjust as
Unit 8
By assessing the distribution of specific markers in all the affected individuals in a population, geneticists can identify DNA markers tightly associated with the disease, thus localizing the disease-associated gene to a relatively small region.
The gene causing diastrophic dysplasia in a large Finnish population was localized to a 60 kb region by this approach.
Let us have a look into more details of it.

Everything we’ve talked about in linkage is based on linkage disequilibrium (LD) within a population. LD is also called gametic phase disequilibrium. LD means that particular alleles at two loci occur together more (or less) often than expected by chance: e.g. A–B and a–b. If two loci are in linkage equilibrium, then the probability of allele “A” being paired with allele “B” equals the probability it is paired with allele “b.”
The longer a population is randomly mated, the less LD will be present, and detection of linkage will become more difficult–i.e., shorter and shorter segments of chromosomes will remain in disequilibrium.
Mapping in outbreed populations–e.g., a human population–can be done due to persistent linkage disequilibrium that had arisen at some point in the past, most likely as a result of a population bottleneck( Sudden decrease in genotypic and allelic frequency in a population due to environmental problems.; Refer Unit 11, Mechanism chapter, for more information on Population bottleneck) The population is probably not truly random mating and tight linkages will persist even after random mating, allowing the detection of linkage between a gene (e.g., cancer) and a molecular marker.
Because of linkage disequilibrium, we can develop and use genetic maps in plants. The cautionary note, however, is that LD will dissipate under random mating, so markers useful initially in a population may lose their effectiveness if the population is intercrossed several times.
Different types of physical maps vary in their degree of resolution. The lowest- resolution physical map is the chromosomal (sometimes called cytogenetic) map, which is based on the distinctive banding patterns observed by light microscopy of stained chromosomes.
A cDNA map shows the locations of expressed DNA regions (exons) on the chromosomal map.
The more detailed cosmid contig map depicts the order of overlapping DNA fragments spanning the genome.
A Macro restriction map describes the order and distance between enzyme cutting (cleavage) sites.
The highest- resolution physical map is the complete elucidation of the DNA base- pair sequence of each chromosome in the human genome.
Physical maps are described in greater detail below.
In a chromosomal map, genes or other identifiable DNA fragments are assigned to their respective chromosomes, with distances measured in base pairs. These markers can be physically associated with particular bands (identified by cytogenetic staining) primarily by in situ hybridization, a technique that involves tagging the DNA marker with an observable label (e.g., one that fluoresces or is radioactive)
The location of the labeled probe can be detected after it binds to its complementary DNA strand in an intact chromosome.
As with genetic linkage mapping, chromosomal mapping can be used to locate genetic markers defined by traits observable only in whole organisms.
Because chromosomal maps are based on estimates of physical distance, they are considered to be physical maps. The number of base pairs within a band can only be estimated.
Until recently, even the best chromosomal maps could be used to locate a DNA fragment only to a region of about 10 Mb, the size of a typical band seen on a chromosome.
Improvements in fluorescence in situ hybridization (FISH) methods allow orientation of DNA sequences that lie as close as 2 to 5 Mb. Modifications to in situ hybridization methods, using chromosomes at a stage in cell division (interphase) when they are less compact, increase map resolution to around 100,000 bp. Further banding refinement might allow chromosomal bands to be associated with specific amplified DNA fragments, an improvement that could be useful in analyzing observable physical traits associated with chromosomal abnormalities.
A cDNA map shows the positions of expressed DNA regions (exons) relative to particular chromosomal regions or bands.
(Expressed DNA regions are those transcribed into mRNA.) cDNA is synthesized in the laboratory using the mRNA molecule as a template; base pairing rules are followed (i.e., an A on the mRNA molecule will pair with a T on the new DNA strand). This cDNA can then be mapped to genomic regions.Because they represent expressed genomic regions, cDNAs are thought to identify the parts of the genome with the most biological and medical significance.
A cDNA map can provide the chromosomal location for genes whose functions are currently unknown.
For disease genes to test when the approximate location of a disease gene has been mapped by genetic linkage techniques.
The two current approaches to high (producing a macro restriction map) and (resulting in a contig map). With either strategy (described below) the maps represent ordered sets of DNA fragments that are generated by cutting genomic DNA with restriction enzymes.
The fragments are then amplified by cloning or by polymerase chain reaction (PCR) methods (see DNA Amplification below).
Electrophoretic techniques are used to separate the fragments according to size into different bands, which can be visualized by direct DNA staining or by hybridization with DNA pr above] The use of purified chromosomes separated either by flow sorting from human cell lines or in hybrid cell lines allows a single chromosome to be mapped (see Separating Chromosomes below). A number of strategies can be used to reconstruct the original order of the DNA fragments in the genome. Many approaches make use of the ability of single strands of DNA and/or RNA to hybridize to form double stranded segments by hydrogen bonding between complementary bases. The extent of sequence homology between the two strands can be inferred from the length of the double Fingerprinting uses restriction map data to determine which fragments have a specific sequence (fingerprint) in common and therefore overlap.

In top down mapping, a single chromosome is cut (with rare cutter restriction enzymes) into large pieces, which are ordered and subdivided; the smaller pieces are then mapped further. The resulting macro- restriction maps depict the order of and distance between sites at which rare cutter enzymes cleave.
This approach yields maps with more continuity and fewer gaps between fragments than contig maps, but map resolution is lower and may not be useful in finding particular genes; in addition, this strategy generally does not produce long stretches of mapped sites. Currently, this approach allows DNA pieces to be located in regions measuring about 100,000 bp to 1 Mb.
The development of pulsed field gel (PFG) Electrophoretic methods has improved the mapping and cloning of large DNA molecules.
While conventional Gel Electrophoretic methods separate pieces less than 40 kb (1 kb = 1000 bases) in size, PFG separates molecules up to 10 Mb, allowing the application of both conventional and new mapping methods to larger genomic regions.
Contig maps: Bottom- up mapping
The bottom up approach involves cutting the chromosome into small pieces, each of which is cloned and ordered. The ordered fragments form contiguous DNA blocks (contigs). Currently, the resulting library of clones varies in size from 10,000 bp to 1 Mb An advantage of this approach is the accessibility of these stable clones to other researchers. Contig construction can be verified by FISH, which localizes cosmids to specific regions within chromosomal bands.
Contig maps thus consist of a linked library of small overlapping clones representing a complete chromosomal segment. While useful for finding genes localized to a small area (under 2 Mb), contig maps are difficult to extend over large stretches of a chromosome because all regions are not clonable.
DNA probe techniques can be used to fill in the gaps, but they are time consuming Technological improvements now make possible the cloning of large DNA pieces, using artificially constructed chromosome vectors that carry human DNA fragments as large as 1 Mb. These vectors are maintained in yeast cells as artificial chromosomes (YACs). (For more explanation, see DNA Amplification below) Before YACs were developed, the largest cloning vectors (cosmids) carried inserts of only 20 to 40 kb. YAC methodology drastically reduces the number of clones to be ordered; many YACs span entire human genes. A more detailed map of a large YAC insert can be produced by subcloning, a process in which fragments of the original insert are cloned into smaller- insert vectors. Because some YAC regions are unstable, large- capacity bacterial vectors(i.e., those that can accommodate large inserts) are also being developed.

The gene mapping is very extensive technique requiring numerous experiments and interpretations begin any research, we got to find out whether the disease/ phenotype genetic. If it is genetic, then it sho be coded by a particular gene or group of genes. To determine its genetic basis, analysis or test crosses. Here we come to know about the genetic inheritance pattern. Now second step is to locate the genes on chromosomes. To localize the genes to particular chromosome, we use very simple, yet efficient technique called Somatic cell hybridization. The technique of somatic cell hybridization is extensively used in human genome mapping. A virus called the Sendai virus has a useful property that makes the mapping technique possible. Sendai virus will be treated with UV rays, so it can no longer cause any infection to the cells. Sendai virus has many sites of attachments. So it can bind to the cells and help in its attachments. Each Sendai virus has several points of attachment, so it can simultaneously attach to two different cells if they happen to be close together. Sendai virus is very small compared to the cell. Hence the cells attached by Sendai virus, are virtually in close
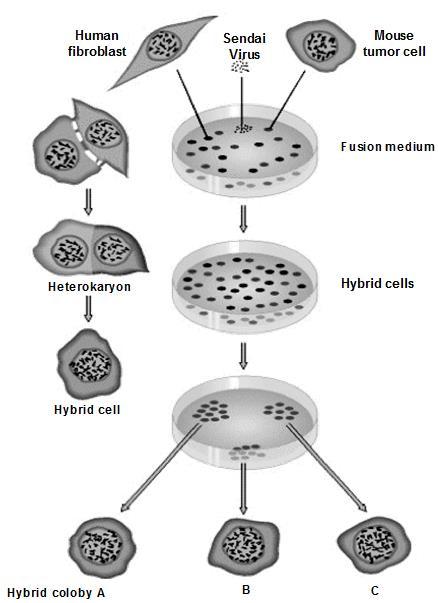
Unit
contact with each other. When cell come too close together, the cell membrane of both cells fuse forming hybrid heterokaryon. Fused cells are called as Binucleate heterokaryone , figure 8.34
If suspensions of human and mouse cells are mixed together in the presence of Sendai virus the virus can mediate fusion of the cells from the different species.
When the cells have fused, the nuclei also fuse to form a uni-nucleate cell is composed of both human and mouse chromosomes.
The mouse and human chromosomes are different in number and shape and hence can be detected very easily.
However, in the course of subsequent cell divisions, for unknown reasons the human chromosomes are gradually eliminated from the hybrid at random.
We can easily determine which human chromosomes have been retained.
Typically, the mouse cell line is mutant for a specific function.
We prepare selective media, which selects only hybrids with human chromosomes.
Thus only those cells will survive, which have got complement for the mouse mutant gene.
We use a DNA marker to locate the specific chromosome, which has survived in the hybrid cells.
The cells are subjected for protein profiling and the protein content is determined

Let us analyze the same with the help
We have 3
Markers are also found in each hybrids. +indicates the presence of markers
-
The presence of certain chromosome in the hybrid cell is giving the phenotype. The absence of one particular chromosome does not result in the desired protein profile. That means that chromosome has the gene of interest. indicatesthe absence ofmarkers. Observe the data carefully We findthatthe Marker A ispresentin only hybrid1 and 2.Hydrid 1 and2 have chromosome 1. at meansmarkerA is on the chromosome 1.Usethesimilar logicforrestmarkers. Markers B chromosome 5. This ishow; we can map manygenes on the human chromosome, usingsomatic cell hybridization.
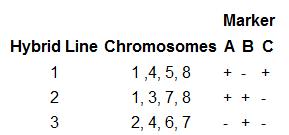
Plantbreeding andgenetics relyon geneticmaps. Theseare madeby analysis ofsegregating plant populations derived by crossing parents with contrasting characteristics.
Solution:
The gene productis shown in only 4 celllines; A,B,D and F
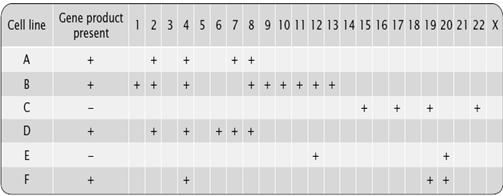
That means all 4 have one chromosomeincommonwhich is giving themthe gene product.
Can youfind out?.....chromosome4is commonto 4of them
Thatmeanschromosome4has thegene.
The hybrid resulting from such a cross is then allowed to produce offspring by selfing, and it is in this segregating family that genetic variation, causing different physical attributes (known as the phenotype), can be assessed.
First molecular marker map: humans 1980
Early plant mapping: tomato, corn and tomato, rice 1988
Examples of saturated maps: tomato and potato (1992), pepper (1999), maize (1999), rice (1998)
First, make a mapping population:
New mutant: tb1-like. This mutant in maize inbred B73 strain.
In figure 8.35, we can see that the population will segregate 3:1for wild type and tb-like plants. You will isolate DNA from some number of mutant (tb) plants.
Every chromosome transmitted is potentially recombinant.

Every F2 mutant can be scored for 0/2, 1/2, or 2/2 recombinant chromosomes.
For each molecular marker to be tested for linkage with “tb” mutant, identify a polymorphism between the B73 and MO17.
Then score inheritance of the polymorphism.
If a molecular marker is very closely linked to the mutation (tb), then those progeny homozygous for the mutation will also be homozygous for the B73 polymorphism
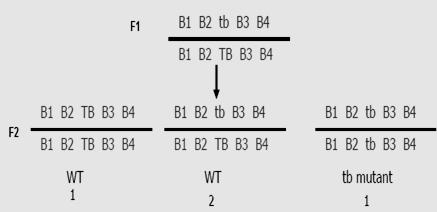
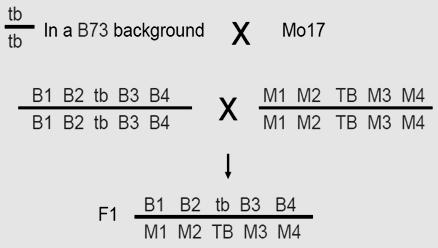
If a molecular marker is unlinked to the mutation (tb), then those progeny homozygous for the mutation are just as likely to inherit the MO17 polymorphism as the B73 polymorphism
If a molecular marker is linked to the mutation (tb), then those progeny homozygous for the mutation are more likely to inherit the B73 polymorphism then the MO17 is polymorphism
F2, backcross, and recombinant inbred are the three primary types of mapping populations used
for molecular mapping. An F oped by selfing (or intermating for cross pollinated species) among F individuals are developed by crossing two parents that show significant polymorphism for whichever type of loci you are going to score. Backcross populations are developed by crossing the F1 with one of the two parents used in the initial cross. The major drawback to using F2 or backcross populations is that the populations are not eternal. Therefore, your source of tissue to isolate DNA or protein will be exhausted at some point in time. You then would have to begin mapping again in another population.
Figure8.35 D: 20 recombinantchromosomes outof a total of 40 scored.Thismarker is unlinked to the mutation
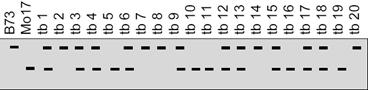

Figure8.35 E:8recombinantchromosomes outof atotal of40 scored 8/40 =0.2 or 20%recombination or 20 mapunits(20 cM between

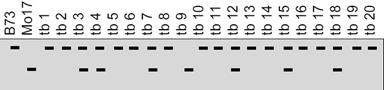
Populations of recombinant inbred lines can be a powerful solution to this problem. Recombinant inbred lines are developed by single-seed selections from individual plants of an F2 population. (Because of this procedure, these lines are also called F2-derived lines.) Single-seed descent is repeated for several generations. At this point, all of the seed from an individual plant is bulked. For example, a F3:4RI population underwent single-seed descent through the F3generation, and was bulked to develop the F4. This population of seed can then be grown to obtain a large quantity of seed of each individual line. Importantly, each of the lines is fixed for many recombination events.
These lines have several uses. First, they can be used be used to derive a map because it essentially is an eternal F population with unlimited mapping possibilities. Additionally, these lines can be scored for morphological traits (such as disease resistance or flower color) or quantitative traits (such as yield or maturity). This morphological trait data can then be compiled and those traits can be placed on the developing molecular map. These lines are especially powerful for analyzing quantitative traits because replicated trials can be analyzed using identical ge- RI population level of in-% within-line homozygosity at netic material. The quantitative trait data can then be used to determine if any molecular markers are closely associated with those traits.
The first type of linkage analysis is the modern cou veloped by Morgan and The method is based on analysis of the progeny of experimental crosses set up between parents of known genotypes and is, at least in theory, applicable to all eukaryotes. Ethical considerations preclude this approach in humans, and practical problems such as the length of the gestation period and the time taken for the newborn to reach maturity (and hence to participate in subsequent crosses) limit the effectiveness of the method with some anima The key to gene to determine the genotypes of the gametes resulting from meiosis. In a few situations this is possible by directly examining the gametes. For example, the gametes produced by some microbial eukaryotes, , can be grown into colonies of haploid cells, whose genotypes can be determined by biochemical tests. Direct genotyping of gametes is also possible with higher eukaryotes if arried out with the DNA from individual spermatozoa, enabling RFLPs, SSLPs and SNPs to be typed. Unfortunately, sperm typing is laborious. Routine linkage analysis with higher eukaryotes is therefore carried out not by examining the gametes directly but by determining the genotypes of the diploid progeny that result from fusion of two gametes, one from each of a pair of parents. In other words, a genetic cross is performed. The complication with a genetic cross is that the resulting diploid progeny are the product not of one meiosis but of two (one in each parent), and in most organisms crossover events are equally likely to occur during production of the male and female gametes. Somehow we have to be able to disentangle from the genotypes of the diploid progeny the crossover events that occurred in each of these two meioses. This means that the cross has to be set up with care. The standard procedure is to use a test cross.
Scenario 1, where we have set up a test cross to map the two genes we met earlier: gene A (alleles A and a) and gene B (alleles B and b), both on chromosome 2 of the fruit fly. The critical feature of a test cross is the genotypes of the two parents: One parent is a double heterozygote This means that all four alleles are present in this parent: its genotype is AB/ab. This notation indicates that one pair of the homologous chromosomes has alleles ‘A’ and the other has Double heterozygotes can be obtained by crossing two purebreeding strains, for example AB/AB The second parent is a pure breeding homozygote parent both homologous copies of chromosome 2 are the same: in the example shown in Scenario 1 both hav alleles ‘a’ the genotype of the parent is ‘ab/ab Our objective is therefore to infer the genotypes of the gametes produced by this parent and to calculate the fraction that is recombinants. Note that all the gametes produced by the second parent (the double homozygote) will have the genotype ab regardless of whether they are parental or recombinant gametes. Alleles ‘a’ and ‘b’ are both recessive, so meiosis in this parent is, in effect, invisible when the genotypes of the progeny are examined. This means that, as shown in Scenario 1 in figure 8.36 A, the genotypes of the diploid progeny can be unambiguously converted into the genotypes of the gametes from the double heterozygous parent. The test cross therefore enables us to make a direct examination of a single meiosis and hence to calculate a recombination frequency and map distance for the two genes being studied.

a, B and b. examiningtheirphenotypes.Becausethe and b - it theprogeny. The thereforethesame as the thatindividual. In Scenario2,AandBareDNAmarkerswhoseallelepairsarecodominant. In this Ab/Ab The individualaredirectlydetected,forexample by PCR. theParent1gametethatgave rise to each individual to be deduced.

Unit
Just one additional point needs to be considered. If, as in Scenario 1, gene markers displaying dominance and recessiveness are used, then the double homozygous parent must have alleles for the two recessive phenotypes; however, if co dominant DNA markers are used, then the double homozygous parent can have any combination of homozygous alleles (i.e. AB/AB, Ab/Ab, aB/aB and ab/ab). Scenario 2 in the figure shows the reason for this.
The power of linkage analysis is enhanced if more than two markers are followed in a single cross. Not only does this generate recombination frequencies more quickly, but it also enables the relative order of markers on a chromosome to be determined by simple inspection of the data. This is because two recombination events are required to unlink the central marker from the two outer markers in a series of three. Either of the two outer markers can be unlinked by just a single recombina-
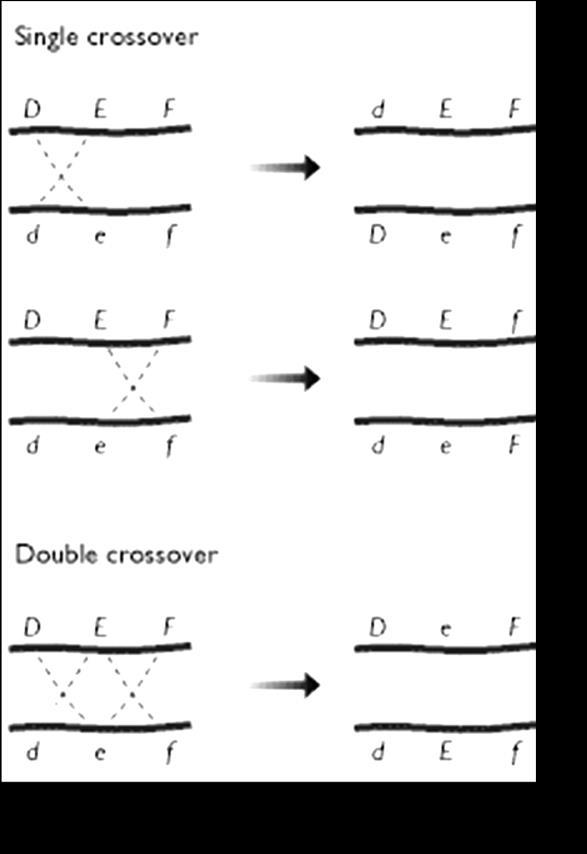
tion.
A double recombinationis less likelythan a single one,so unlinkingofthecentral marker willoccurrelatively infrequently.
An example ofthe data obtained froma three-point cross isshown in the table. A cross hasbeen set up between a triple heterozygote ( ). The mostfrequent progenyare thosewith one ofthe two parental genotypes,resultingfroman absence ofrecombinationevents in the region containing the markers A, B andC.Two other classes of progenyare relativelyfrequent (51 and 63 progeny in the example shown). Eachofthese is presumed to arise from a single recombination. Inspection oftheir genotypes shows thatin the first ofthese two classes, marker A hasbecome unlinked fromB and C, and in the secondclass,markerB has becomeunlinked fromA andC.
The implication is that A and B are the outer markers. This is confirmed by the number of progeny in which marker C has become unlinked from A and B. There are only two of these, showing that a double recombination is needed to produce this genotype. Marker C is therefore between A and B.
Inferredrecombination
None (parental genotypes)
Aand B + C
One, between Band A + C
and A and one and B
16.Howwouldyoupredictthewhetheracharacter is actuallyageneticallycoded or environmentallycoded
17.Linkedgenesalwaysstayliked. Is thestatementtrue? If yes/Nowhat is yourexplanation?

18.Linkageandrecombinationaretheproperty of genesandchromosomes,does it depend on presence or absence of introns?
19. How manylinkagegroups,humanmalehas?
20 If therewerecompletelinkagebetweenMendeltallanddwarfgenes,howtheF2ratio wouldappear?
Mitochondrial and Chloroplast contain chromosomes which are involved in Extra chromosomal inheritance
The Chromosome of both the organelle is circular and resembles the prokaryotic DNA

is believed that, these organisms were once free living and started mutual relationship with the eukaryotic cells, as stated by Endosymbiotic theory
female egg cell provides Cytoplasmic contents to the zygote and along with cytoplasm , organelle chromosomes also get inherited
Development of Cytoplasmic sterile plants has been a major breakthrough in plant breeding technology
In human, the resistance to chloramphenicol antibiotic is inherited maternally

Maternal inheritance Is different form maternal influence

Maternal inheritance stands for the inheritance of organelle chromosomes through cytoplasm of egg
Maternal influence stands for the influence of maternal Cytoplasmic contents such as RNA s and proteins during the initial zygote formation and development.

We are aware that some organelles in the eukaryotes contain genetic material. It is not the only property of nucleus. Other major organelle, Mitochondria and Chloroplast possess their own genetic material. These genetics material code for several proteins and enzymes, influencing many characters along with nuclear genes. In this section we shall discuss about the genes present in Cytoplasmic organelles and their influence on inheritance.
Criteria for extra nuclear inheritance
1. Differences in result of reciprocal crosses
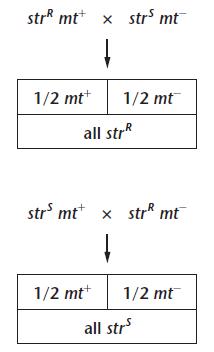
2. Genes are getting inherited from only one parent.
3. Failure to locate genes on nuclear chromosomes. That means they are on the Cytoplasmic chromosomes.
4. Lack of Mendelian segregation and lack of Mendelian ratios
5. Transmission of traits, even though nucleus has been removed by genetic engineering.
Two organelles in the eukaryotic system contain genetic material roplast and Mitochondria. Inheritance of genes from these organelles is termed as Organelle heredity.
The organelles possessing genetic material were once free living organisms In the course of evolution, they developed symbiotic relation with eukaryotic cells. Thus they started living inside the eukaryotic cells, becoming a part of the cell.
Mitochondria were once the free living bacteria
Chloroplast was once free living algae.
These brought with them DNA and other protein mechanisms and started living inside the cell. Mitochondria provide us with ATP and we give it protection and other necessary chemicals. Chloroplast provides us with Energy and Carbohydrates; in return we protect the chloroplast and provide nutrition.
Chloroplast mutations in Chlamydomonas is unicellular green algae. They are haploid in nature and possess single larg of circular DNA molecule
Chlamydomonas has 2 mating types : mt equally to the zygote and hence their nuclear chromosomes donate equal no. of genes. But crosses, have shown that mt the strepto
The inheritance of resistance to streptomycin is uniparental in Chlamydomonas.

Olderaretheparents,morechance of genetic abnormality in kids

result of different.This indicatestheinfluence of mother morethanthefather
Figure8.38 The streptomycinresistance is passedonly by mt+strain. Even if the mt-strainhas thestrRgene, itis not transferred to theprogeny
Chlamydominas,a eukaryotic green alga, may be sensitive to the antibiotic erythromycin, which inhibits protein synthesis in prokaryotes. There are two mating types on this alga, mt+ and mt-. If a my+ cell sensitive to the antibiotic is crossed with a mt-cell that is resistance, all progeny are sensitive. The reciprocal cross ( mt+ resistant and mt sensitive) yields all the resistance progeny cells. Assuming that the mutation for resistance is in the chloroplast DNA, what can you conclude from the results of these crosses?
HINT:
This problem involves an understanding of the Cytoplasmic transmission of organelle in unicellular algae. The key to its solution is to consider the results you would expect from two possibilities: that inheritance of the trait is uniparental or that inheritance is biparental
8.5.1.1 Plastid inheritance in Mirabilis

In Mirabilis Jalapa, threetypes of branches occur. They are green, variegated and colorless. Green branches contain green plastids in their leaves Variegated branches contain green plastids and colorless plastids. So the leaves have white spots. Colorless branches are due to the presence of colorless plastids. Seeds collected from green branches produce only green plants. Seeds collected from white branches produce only white plants (which cannot survive because of the absence of chlorophyll). Seeds collected from variegated branches produce three kinds of plants, namely green, variegated and colorless
When flowers on a green branch are pollinated with pollen from a colorless branch, all the resulting offspring are green
When flowers on a colorless branch are pollinated by pollen from a green branch, all the offspring are colorless
When flowers on variegated branch are pollinated by pollens from any branch, all the three types of offspring are produced.
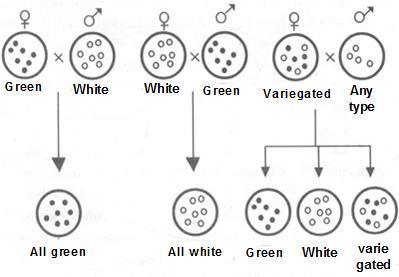
These experiments show that the phenotype of the off-springs is determined by the parent who contributed the egg. This is because the offspring receive cytoplasm with plastids only through the egg. The gene conferring the chloroplast and its property is mainly located in the chloroplast genes. Thus this experiment also shows that the chloroplast codes for its own proteins and other structural materials.
Mitochondrial DNA (mtDNA) is the DNA located in organelles called mitochondria, structures within eukaryotic cells that convert the chemical energy from food into a form that cells can use, ATP. Most other DNA present in eukaryotic organisms is found in the cell nucleus.
Unit 8
Some of the estimated 20,000 genes in the human genome are located in small compartments in the cell called the mitochondria, rather than on chromosomes in the cell’s nucleus. Some cells contain many hundreds of mitochondria The genes found within the mitochondria contain the information that codes for the production of many of the important enzymes that drive the biochemical reactions to produce the body’s source of energy: a chemical called ATP (adenosine triphosphate). The cells in the body, especially in organs such as the brain, heart, muscle, kidneys and liver, cannot function normally unless they are receiving a constant supply of energy.
Faulty mitochondrial genes can result in absence of these enzymes, or enzymes that are impaired and do not work properly. This leads to a reduction in the supply of ATP, and may result in problems with the body’s functions The pattern of inheritance of conditions due to faulty mitochondrial genes is often called maternal inheritance. This is because a child inherits the great majority of their mitochondria from their mother through the egg. Usually a mother will have a mixture of mitochondria containing the working gene copy and others containing the faulty gene. For a condition to develop, the number of mitochondria with the faulty gene must be above a critical level (the threshold)
The cells of different tissues and organs can have varying amounts of mitochondria with a faulty gene, and the number of cells with the faulty mitochondria in each tissue or organ may vary Conditions due to having faulty mitochondrial genes create complexities when assessing the risk the mother has for passing on the faulty mitochondrial genes to a child and if, or how severely, the child would be affected
While it is possible to test for the presence or absence of some faulty mitochondrial genes and their products during pregnancy, the result may be difficult to interpret Whether symptoms of the condition will occur or not depends on whether the numbers of mitochondria with the faulty genes is above the critical threshold in enough cells to interfere with energy production. Thus number of defective mitochondria is critical to inherit the faulty genes.
Table 8.40. Showing the different protein complexes coded byMitochondrial genes. Abovefigure 8.40 shows the location and arrangement of these genes on the mitochondrial chromosome
ND1, ND2, ND3, ND4, ND4L, ND5, and ND6-of oxidoreductase)
HumanmtDNAencodes 13 polypeptides: 7 subunits
•1 subunit-cytochrome b -of Complex III (CoQ-cytochrome c oxidoreductase)
•3 subunits-COX I, COX II, and COX III -of Complex IV (cytochrome c oxidase, or COX),
•2 subunits-ATPase6 and ATPase 8 -of Complex V (ATPsynthase).
•Manymore believed functions and subunits , but not yet confirmed experimentally
It also encodes 2rRNAsand 22tRNAs,required for its own expressions

Genetic counseling can provide the most current information on the availability and appropriateness of testing for mitochondrial conditions, either in an affected person or during pregnancy.

Mitochondria is easy target for any mutations, because
1. It has no Histone proteins which protect the DNA.
2. Mitochondria concentrate highly mutagenic reactive oxygen species (ROS), produced by cellular respiration.
Human mitochondria has 37 genes coding various proteins. Human cells contain 100s of mtDNA in somatic cell Human oocyte contains 100,000 copies of mtDNA Thus if, few of them have mutated, it will not have any direct effects on the cell. Due to presence of large number of normal mtDNA, mutant effect gets diluted. In humans the mitochondrial inheritance has following characteristics:
1. Inheritance exhibits maternal pattern, and no Mendelian patterns
2. Disorder shows a deficiency in the organelles function
3. There will be mutation in one or more of the mtDNA genes. Figure 8.41 shows the pattern of mitochondrial inheritance.
In Chlamydomonas, the mitochondrial genome lacks two important genes, cox2 and cox3, which are instead encoded by the nuclear genome. These genes contain multiple eukaryotic spliceosomal introns as well as a mitochondrial targeting sequence and could therefore represent the original, premitochondrial genes that were later transferred to the mitochondria. A subunit of the ATPase in Chlamydomonas is also nuclearencoded and contains spliceosomal introns and a mitochondrial target sequence By comparing nuclear-encoded cytoplasmic ribosomal proteins (CRPs) and mitochondrial ribosomal proteins (MRPs), it was show that both proteins contained conserved intron positions, indicative for their common descent and eukaryotic origin. The CRPs and MRPs amongst themselves share also many intron positions, showing that the mitochondrial proteins that are nuclear-encoded show a common origin between all eukaryotes
Mother is compared to the GOD and she is the initial teacher and the best guardian. She molds child’s initial
development and character building. It is also true from genes point of view. We learn that mother and father donate equal number of genes to the offspring. But mother’s influence on child is more than father, as she donates cytoplasm in egg cell, which father fails to deliver. The Cytoplasmic organelle, contribute their gene, which is called as Mitochondrial Inheritance. Along with the organelles, the mother cytoplasm also donates important proteins and
RNA molecules for the zygote and embryo development. Thus the mother influences the child growth by her Cytoplasmic components. This is not inherited but only possessed by the child. The best studied example is the sail coiling pattern, which is governed by Cytoplasmic components of mother. Shell coiling in snail Limnaea peregra is an excellent case of maternal effect. Some strains of snail have left handed shell coiling; and some have right handed coiling. Right handed = Dextral Left handed = sinistral
The Coiling of the shell is maternally influenced. The genes ’D’ causes the Right coiling, as it is Dominant. The recessive‘d’ causes the left handed coil, as it is recessive condition. The coiling depends on the maternal genotype more than the off spring genotype. Any offspring, whose mother has one copy of ‘D’ gene will be right handed.

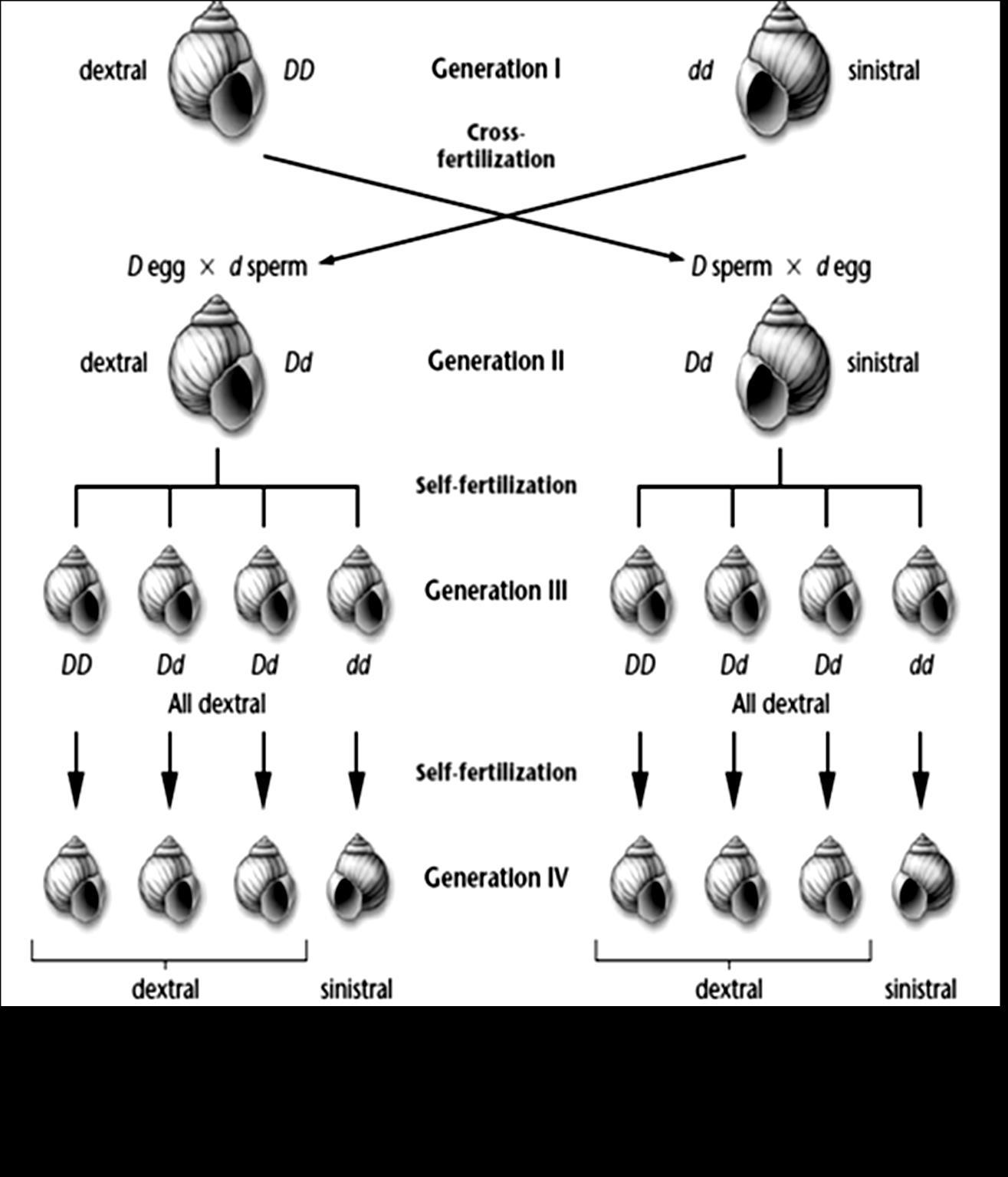

What can we conclude form the diagram above?
The coiling pattern is not dependent only on the genotype of the offspring
The coiling pattern of the progeny is determined by the genotype of the female snail, regardless of the phenotype of the female
Females that are either ‘DD’ or ‘Dd’ produce oocyte with D gene product.

if the fertilized egg contains only dd alleles, it will show dextral coiling. Molecular factor in shell coiling
Orientation of spindle fibers in the first cleavage after fertilization determines the direction of coiling
Spindle orientation is controlled by maternal gene products, present in the oocyt
This establishes permanent coiling pattern
D allele, causes dextral coiling
d allele, causes sinistral coiling
Thus the maternal influence has immense role in shaping the snail coils. Along with genetic imprinting, maternal influence is another powerful phenomenon, which
23 If maternalinheritanceandmaternaleffect has intenseinfluence on theorganism,then why don’t we allresembleourmother?
do youthink when might be themRNAforeggformation in females is produced in femalebody?
25.Whatwould be thereasonforseveresideeffects in ourbodyunderoverdose of antibacterialmedicine.


Microbial genetics is the most basic and very important part of genetics. We use microbes as the model organism for many genetic studies and genetic engineering. Hence locating and analyzing the genes on the microbial DNA is very important
Transformation- it is the uptake of naked ssDNA by any competing bacteria. The up taken DNA undergoes homologous recombination with the bacterial genome. Thus it provides with new set of alleles and makes the bacteria recombinant.

Transduction – the transfer of genetic elements is via bacteriophage. Bacteriophage infects the bacteria and during lytic stage, in can take up some of the bacterial genes also. During second infection cycle to another bacterium, the recombination takes place.
Conjugation – the conjugation is the process of process of acquiring new genes and alleles from other live bacteria via a conjugation tube. Hfr strains have high frequency of transferring the genes to other bacteria. The bacteria which can form a conjugation tube are called as ‘F+’ bacteria and the one which cannot form the conjugation tube is called ‘F-‘ bacteria
Genes on the bacterial chromosome are mapped using Interrupted mating experiments.
We are outnumbered 10:1 by bacteria on our bodies.

Nearly8% of humangenome is derivedformretro transpositionthroughoutevolution
Microbes are ubiquitous in nature. We can never find a place without a single microorganism. In fact, evolutionary biologists have strongly claimed that, Microbial world was the primitive life on earth followed by multi cellular organisms. In this section, we shall focus on microbial genetics, their mode of replication, mode of gene transfer and techniques to map their genome.
Bacteria are the simplest microbes, and our very reference of microbe means bacteria. They are very simple organisms and have smaller genetic content. Let us understand the basic characters of bacteria:
They have simple circular DNA as genetic material
They have no nucleus and devoid of any membrane bound organelle
They possess, extra nuclear DNA called, Plasmids Plasmids are circular, self-replicating DNA bodies which exist independently in the bacterial cytoplasm The plasmid can integrate into bacterial chromosome, and sometime it can come out and live freely, called Episome.

Bacteria are very famous for their asexual, binary cell division. In 1946, Lederberg and Tatum discovered new ways of gene transfer between 2 cells.
1. Conjugation
2. Transformation
3. Transduction
All bacteria do not exhibit all types of gene transfer methods. Transformation occurs in limited cases in bacteria Conjugation is the most preferred choice by most bacteria Thus, bacteria do reproduce sexually just like higher living beings. Each type of these methods consists of one way movement of genetic information to the recipient cell. It may / may not be followed by recombination These processes are not connected to cellular reproduction in bacteria The cellular reproduction involves creation of new cells But these gene transfers does not create a new cell in the colony, it just converts one type of bacteria into other.
Q: Is sexual reproduction of bacteria is same as higher organism? No, it differs in 2 aspects.
1.DNA exchange and reproduction are not coupled in bacteria
2.Donated genetic material that is not recombined into the host DNA is usually degraded. Thus recipient remains haploid.
Insertion of foreign DNA requires homologous recombination. The bacterial system needs some special proteins for homologous recombination called Rec proteins Rec A protein plays major role in ssDNA recombination. Rec A is the most common type of enzyme required for insertion of foreign DNA. Rec BCD proteins, consists of polypeptides coded by 3 rec genes required for dsDNA recombination.
Conjugation involves the transfer of genetic material from one bacterium to another. We call them as Donor and recipient. As you can see in the diagram, a Cytoplasmic bridge is formed between 2 cells and DNA gets transferred ( called as Sex pili or conjugation tube) The homologous recombination occurs between the new incoming DNA and recipient chromosome and the incoming DNA gets incorporated into the recipient system. The part which is not recombined, gets degraded by cellular DNAase enzymes F+ and F- cells
Conjugation depends on a fertility factor (F) that is present in the donor cell and absent in recipient cell ‘F’ Factor is a plasmid containing some specific genes. Cells that contain F factor are called F+ Cells that lack F factor are called as F- What is so important about F factor?
It contains Origin of replication
It also contains no. of genes required for conjugation
It encodes proteins necessary to form the sex pili Sex pilus in different from usual bacterial filament, which is much longer than the sex pilus.

The F factor is nicked at an OriT location .One end of the nicked strand separates from the circle and passes into recipient cell. Replication takes place on the nicked strand, following rolling circle mode of replication. The ‘OriT’ site is the first to go in to the recipient cell. Once the DNA is inside the recipient, it is replicated to produce a dsDNA copy of F plasmid. If the entire F factor is transferred to recipient, then it also becomes a F+ cell.
Fig8.45.Showingconjugationbetween
F-strain.Notethatafterconjugation the Fdoes nto
Sometimes, the F plasmid can get integrated inside the donor chromosome, to form an episome. This when passed onto F- cell, can take some bacterial genes along with F plasmid genes. Thus we can say when F plasmid is integrated inside the chromosome, higher chance that bacterial genes are also transferred along with F factor. Such cells we call as Hfr cells, (High frequency of recombination) Hfr cells behave as F+ cells, forming sex pili and undergoing conjugation with F- cells. Never a Hfr can for pilus with F+
Sometimes the integrated F factor, in Hfr may get excise from chromosome. During excision, the F factor may take some chromosomal genes along with it. Cells containing an F plasmid with some bacterial genes are conjugation occurring between F’ and Fcells can transfer some of the bacterial gene to the F- along with F factor. The conjugation between F’ and F- is Sexduction. Sexduction produces partial diploid cells (Merozygotes)
Note:
Conjugation in E. coli is controlled by an Episome called the F factor
When F factor excise from the bacterial chromosome, it may carry some bacterial genes ( in this case lac genes) with it
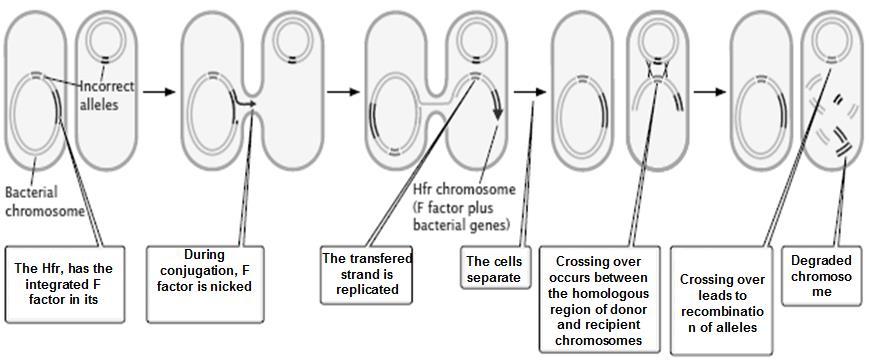

Figure8.46.Diagramshowing the formation ofF’ bacteria.
Cells containing F (F+ cells) are donors during gene transfer; cells without F (F- cells) are recipients.
Hfr cells possess F integrated into the bacterial chromosome; they donate DNA to F- cells at a high frequency.

It is the second method of horizontal mode of gene transfer in bacteria. This mode of gene transfer helped early biologist to conclude DNA is the genetic material. Transformation requires both uptake of DNA from the surrounding and its incorporation into bacterial chromosome or a plasmid. Cells that take up DNA are said to be competent. The competency depends on growth conditions, DNA concentrations and composition of the medium.
DNA uptake is a random event, any DNA can be up taken, need not be only bacterial origin strand can enter the cell and the other strand is degraded. Inside the cell ssDNA undergoes homologous recombination and gets integrates into bacterial chromosome. Non-homologous ssDNA is degraded.
If the foreign DNA has an origin of replication recognized by the host cell DNA polymerases, the bacteria will replicate the foreign DNA along with their own DNA. When transformation is coupled with antibiotic selection techniques, bacteria can types and conjugation conjugation)
be induced to uptake certain DNA molecules, and those bacteria can be selected for that incorporation. Bacteria which are able to uptake
DNA are called "competent" and are made so by treatmentwith calcium chloride in the earlylog phaseof growth. The bacterialcellmembrane is permeable to chloride ions, butis non-permeable to calcium ions.As the chloride ionsenterthe cell, water molecules accompanythe charged particle. Thisinfluxof watercauses the cells to swell and is necessary for the uptake ofDNA.The exactmechanism ofthisuptake is unknown. Itis known, however,that the calcium chloridetreatment be followedby heat.When E.coliare subjected to 42°C heat, a setofgenes are expressed which aid the bacteria insurviving at such temperatures.Thisset ofg enesis called the heat shock genes. The heatshock stepis necessary for the uptake ofDNA.Attemperatures above 42°C, the bacteria's ability to uptake DNA becomes reduced, andat extreme tempe ratures the bacteria will die.
The process for the uptake of naked plasmid and bacteriophage DNA is the same; calcium chloride treatment of bacterial cells produces competent cells which will uptake DNA after a heat shock step. However, there is a slight, but important difference in the procedures for transformation of plasmid DNA and bacteriophage M13 DNA. In the plasmid transformation, after the heat shock step intact plasmid DNA molecules replicate in bacterial host cells. To help the bacterial cells recover from the heat shock, the cells are briefly incubated with non-selective growth media. As the cells recover, plasmid genes are expressed, including those that enable the production of daughter plasmids which will segregate with dividing bacterial cells.

Unit 8
However, due to the low number of bacterial cells which contain the plasmid and the potential for the plasmid not to propogate itself in all daughter cells, it is necessary to select for bacterial cells which contain the plasmid. This is commonly performed with antibiotic selection. E. coli strains such as GM272 are sensitive to common antibiotics such as ampicillin. Plasmids used for the cloning and manipulation of DNA have been engineered to harbor the genes for antibiotic resistance. Thus, if the bacterial transformation is plated onto media containing ampicillin, only bacteria which possess the plasmid DNA will have the ability to metabolize ampicillin and form colonies. In this way, bacterial cells containing plasmid DNA are selected.
The transformation of bacteriophage M13 into bacterial cells is identical to plasmid DNA transformation through the heat shock step. After the heat shock step, single stranded M13 DNA begins replicating in the host cell through use of the host cell machinery. During the life cycle of this virus, however, M13 replicative form is created and daughter phages are packaged and extruded from the bacterial cell. These intact phage molecules then infect neighboring bacteria in a process called transfection. When these transformed and transfected bacteria are plated with non-infected cells onto growth media, the non-infected cells form a background cell lawn which covers the plate. In regions of M13 transfection, areas of slowed growth, called plaques, can be identified as opaque regions which interrupt the lawn.


Since M13 viral transfection is a critical part of the transformation of bacterial cells with M13, it is absolutely necessary to use a strain of E. coli which harbors the episome for the F pilus. When M13 phages infect bacterial cells they attach to the F pilus, and the loss of this pilus is a common reason for a failed or poor transformation/transfection of M13. JM101 is a strain of E. coli which possesses the F pilus if the culture is maintained under appropriate conditions. Since the F pilus is not necessary for plasmid DNA transformation, it is advisable to use GM272, a much healthier, F- strain of E. coli for this procedure. To avoid confusion between the similar procedures, bacterial transformation with plasmid DNA is termed a "Transformation", and a bacterial transformation with naked M13 followed by a transfection with intact M13 phage is called a "Transfection."
The order of gene can be mapped by analyzing the transformation patterns and order of gene uptake. In transformation, the bits and fragment of DNA gets up taken. We can easily find out the gene present in the
Transformation, like conjugation, is used to map bacterial genes, especially in those species that do not undergo conjugation or transduction .Transformation mapping requires two strains of bacteria that differ in several genetic traits; for example, the recipient strain might be a_ b_ c_ (auxotrophic for three nutrients), with the donor cell being prototrophic with alleles a+ b+ c+. DNA from the donor strain is isolated and purified. The recipient strain is treated to increase competency, and DNA from the donor strain is added to the medium. Fragments of the donor DNA enter the recipient cells and undergo recombination with homologous DNA sequences on the bacterial chromosome. Cells that receive genetic material through transformation are called transformants. Genes can be mapped by observing the rate at which two or more genes are transferred together (co-transformed) in transformation. When
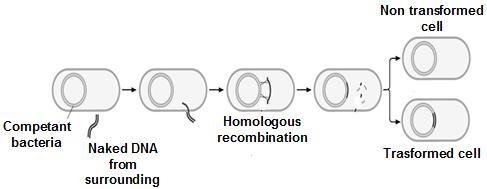
the DNA is fragmented during isolation, genes that are physically close on the chromosome are more likely to be present on the same DNA fragment and transferred together Genes that are far apart are unlikely to be present on the same DNA fragment and rarely will be transferred together. Once inside the cell, DNA becomes incorporated into the bacterial chromosome through recombination. If two genes are close together on the same fragment, any two crossovers are likely to occur on either side of the two genes, allowing both to become part of the recipient chromosome. If the two genes are far apart, there may be one crossover between them, allowing one gene but not the other to recombine with the bacterial chromosome. Thus, two genes are more likely to be transferred together when they are close together on the chromosome, and genes located far apart are rarely co-transformed. Therefore, the frequency of co-transformation can be used to map bacterial genes. If genes a and b are frequently cotransformed, and nes b and c are frequently cotransformed, but genes a and c are rarely cotransformed, then gene b must be between a and c—the gene order is a b c. E.g., if 50 met+ his-, 50 met- his+, and 100 met- met-; then (50+50)/(50+50+100) =.5
This is another type of horizontal gene transfer technique, which involves the bacteriophage infection process.
It was discovered by Zinder and Lederberg in 1952, in Salmonella typhimurium

Virulent phages can reproduce strictly through the lytic cycle and always kill their host cell. Temperate phage can utilize either lytic or lysogenic life cycle. They do not burst the host rapidl, rather gets integrated into the bacterial chromosome. DNA gets integrated inside the bacterial chromosome and remains in inactive state.We call integrated viral gene as Prophage. It replicates along with bacterial genome and multiplies in number. When the bacterial cell is aging and when conditions are not favorable for phage, the prophage excises from the chromosome and enters lytic stage.
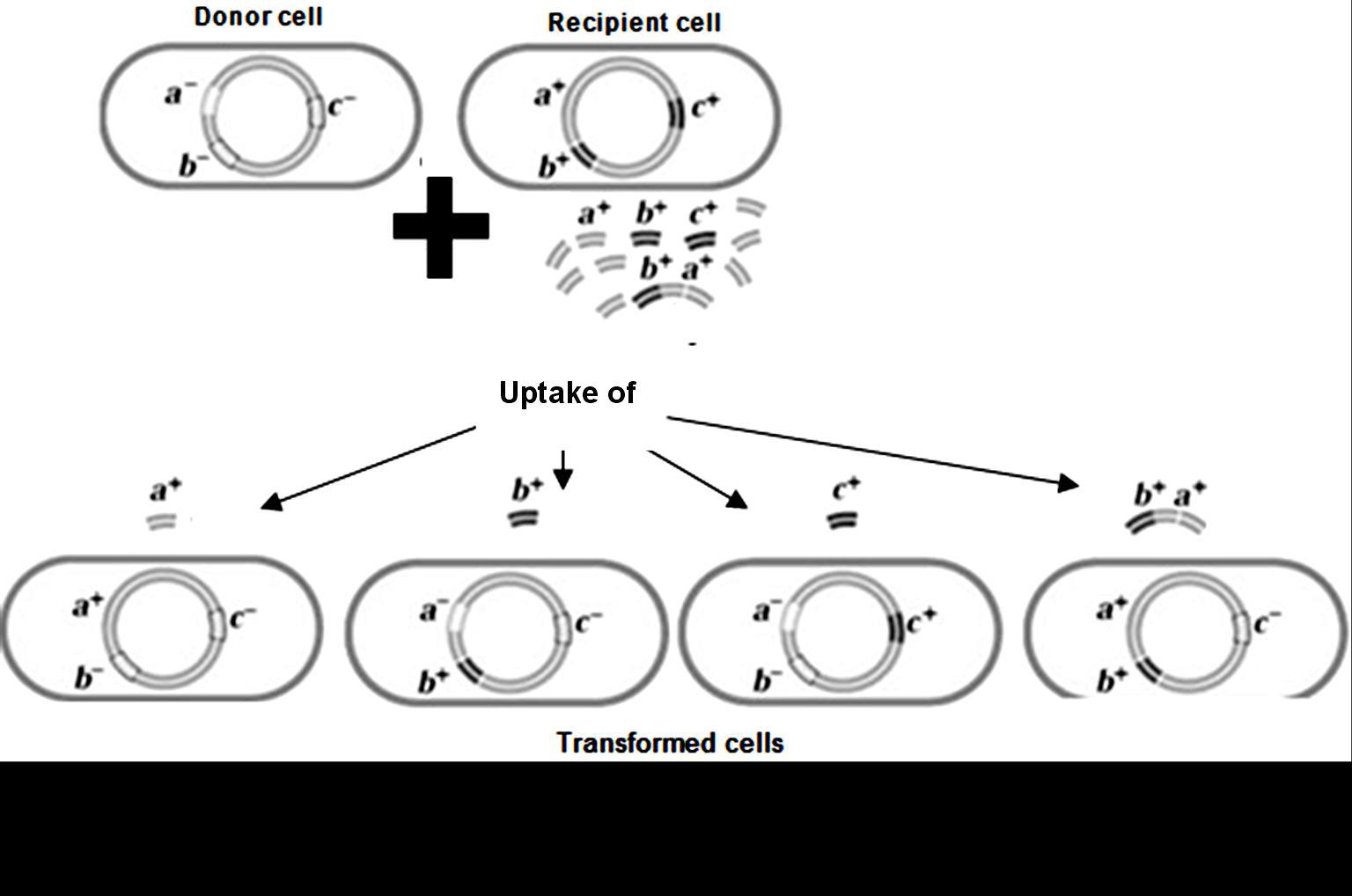
This experiment is used to determine whether cell contact is required for recombination in bacteria. The test helps to determine the mode of gene transfer, by eliminating various possibilities. Let us see this in detail below. The 2 arms of Davis U-tube are separated by a glass filter. (Refer figure 8.50)
Filter has pores which allow only DNA molecules to pass and not the live cells.
of one genotype are placed in one arm
Bacteria of another genotype are placed in the other arm
Lets us use some DNAase in the tube.( DNAase degrades any naked DNA in media)

still recombinants are formed, that means recombinants are not produced by transformation, which requires uptake of naked DNA
Figure8.50.TheDavisU- tube helps to determine the mode of genetransferbetween the bacteria. On two arms of the U-tube, we take2differentstrain of bacteriadeficient in alternatemediarequirements. Onlywhenthey can recombine by exchangingthe gene, we obtainhybridswith all wild(+)typegenes.
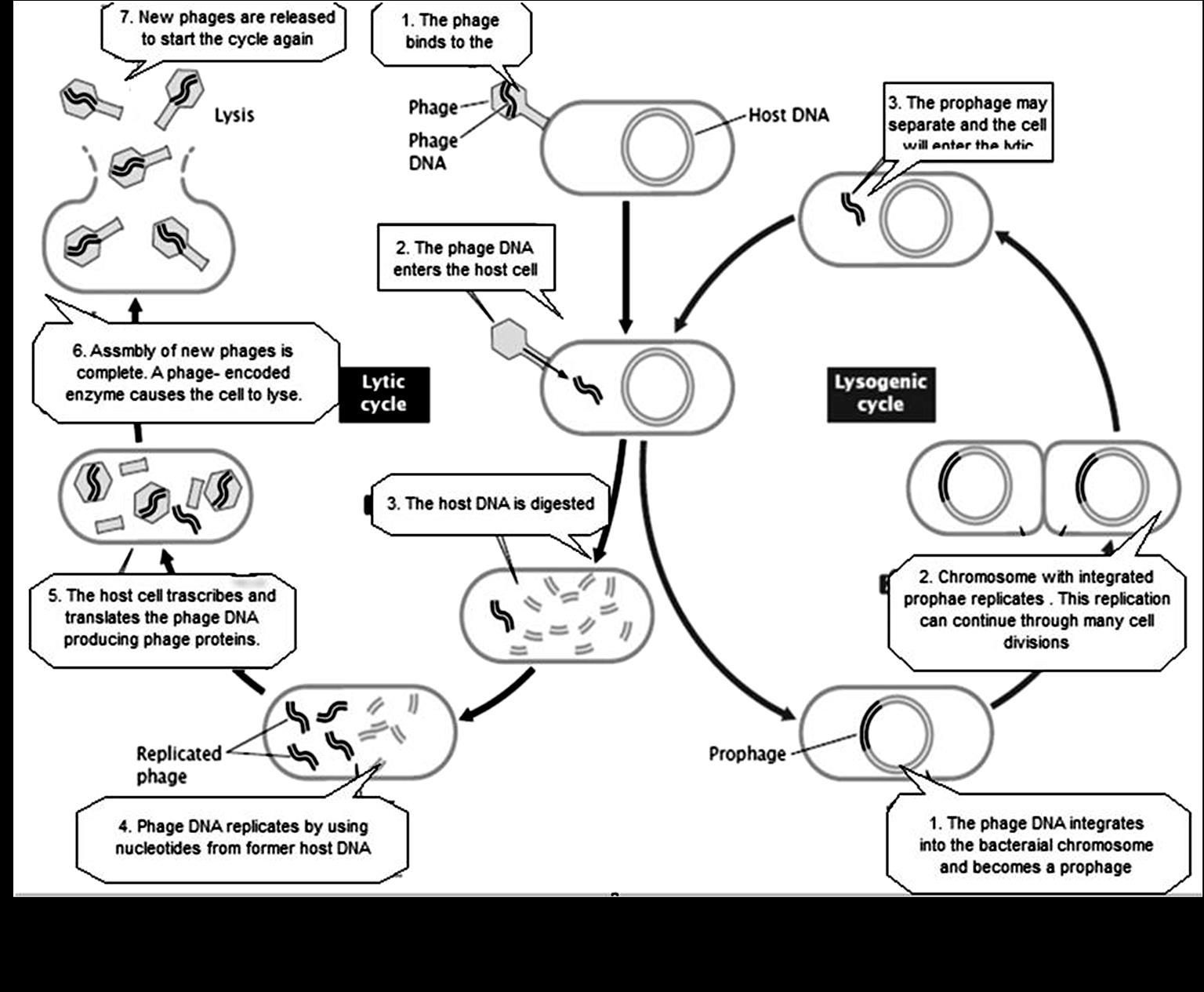
they are separated by
conjugation possible.
still recombinants are occurring,
means
the recombinants are formed due to transduction process, by bacteriophage
the Davis U-tube helps to differentiate the type of recombination process.

on addition of DNAase
not appear, then it must be the Transformation.
recombinants arise even after adding DNAse
must be by transduction or conjugation.
adding the filter which does not allow for cell cell contact, recombination does not occur, means it must have been by conjugation.
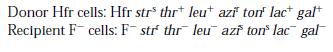
Transfer of entire Hfr needs 100 minutes of conjugation process.
If conjugation is interrupted in between, only some part of the genes are transferred
Genes are always passed in the order of their arrangement on the chromosomes

Distance between 2 genes is expressed in terms of Recombination frequencies or percentages.
Basic unit of distance in conjugation maps
Let us learn the technique with the help of an experiment.
Let us take a donor Hfr is sensitive to antibiotic streptomycin (str ), resistant to sodium azide (azrr) , Resistant to bacteriophage T1 infection (ton Prototrophic for leucine ( leu+) Able to break down lactose ( lac
Let us take a F- recipient is resistant to antibiotic streptomycin (strr), sensitive to sodium azide (azr riophage T1 infection (ton Auxotrophic for leucine ( leu
Let us allow
After few minutes, we shall interrupt the conjugation process.
Unable to break down galactose (gal )
Atregular intervalthe sample cells were removedand agitated vigorouslyin a kitchenblenderto
The genes are passed from Hfr to F- in ordered manner. This helps us to map the genes present in the bacterial chromosome this section we shall study how to map the bacterial genome by interrupted mating experiments. break the conjugation tube.
Plate the cells on a selective media that has streptomycin and lacks leucine and threonine.
The original donor hfr are streptomycin sensitive, hence they will die on the medium
The F- cells were auxotrophic for leucine and threonine , and hence they will also die in the medium
againsttheYchromosomeswhichhasonly 60 million bases
Only the conjugated cells which have received leucine and threonine genes will survive.
Now lets us see, how these genes are arranged.
We shall interrupt the mating process at 9th minute.
We found that it has azrr gene. ( it could grow in medium contain streptomycin and sodium azide)
Then when we stopped at 10th min, the exconjugates had ton
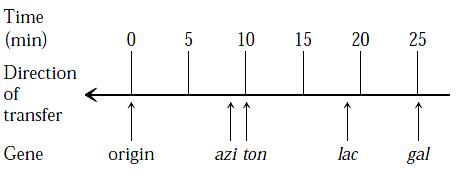
When stopped at 18 had lac
And when stopped at 25 they had gal
These transfer times indicate the distance between the genes on the chromosomes.
More distant the genes, the frequency of Exconjugates decease
Ex: 90% received the azr received gal
Different Hfr strains have the F factor integrated into the bacterial chromosome at different sites and different orientation Gene transfer always begins within F factor. The position of F factor determines the direction of gene transfer

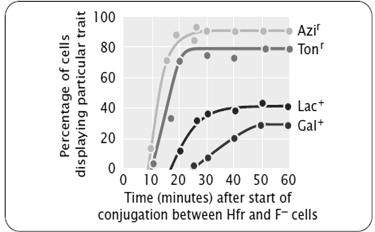
In Hfr 1 the gene transfers continuous in clockwise direction the genes are transferred. Hence the order will be leu-thr-thi-gal-lac-pro-azi Hfr 5 has F integrated between thi and his genes. Transfer occurs in clockwise direction
Interruptedmatingexperiments in gettingtransferredand thebacterialchromosomes. In 8th minuteonlythestr+,leu+andthr+geneshavegone,meansthey tonandthen
time interval between the gene transfers, we can map the genes on the circular chromosome of bacteria.
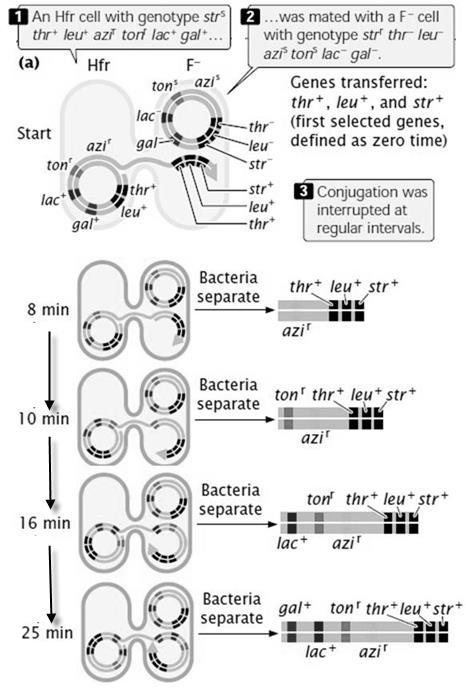


Solved Problem : 1
DNA from a strain of Bacillus subtilis with genotype a+ b+ c+ d+ e+ is used to transform a strain with genotype a-b-c-d-e-. Pairs of genes are checked for co-transformation and the following results are obtained: a+ and b+ no a+ and c+ no a+ and d+ yes a+ and e+ yes b+ and c+ yes b+ and d+ no b+ and e+ c+ and d+ no c+ and e+ yes d+ and e+

On the basis of these results, what is the order of the genes on the bacterial chromosome?
Close located genes, will go together. Hence a+ and d+ are linked, that means they are close located. Use same logic for all. Order will be :
Solved Problem :
An F+ strain was marked at 10 loci, giving rise to hfrprogeny at various integration sites. From the following data determine the order of markers in the F+ strain: Hfrstrain Order of markers donated 1 -Z.H.E.R-> 2 -O.K.S.R -> 3 -K.O.W.I-> 4 -Z.T.I.W -> 5 -H.Z.T.I -> What is the order of the genes? Solution: Order is = Z H E R …look at the below strain, it has R gene and S K O. third one has K O common, followed by W and I ….
Last strain has T I is common and H and Z Thus order will be It is a circular chromosome,.
We use the term gene every now and then, but interestingly there is no typical gene with a specific structure and features. Genes are now known to exist in all possible designs and to function in many different ways. The concept of gene which began almost 100 years ago has seen remarkable changes with every new research pointing out newer facts about gene.
Classical Concept of Gene - Unit of Function, Mutation and Recombination
Unit 8
The term gene was coined by ‘Wilhelm Johannsen’ in 1909. He described it as just as the numbers used in counting with no physical or chemical units of their own.

Thomas Hunt Morgan introduced the tiny fruit fly or Drosophila melanogaster for genetic studies and the rediscovery of Mendel’s laws was a turning point in the young field of genetics The fruit fly has ever since continued t be the young ‘Cinderella’ of genetics. The elegant analysis of inheritance patterns of different traits in Drosophila by Thomas Hunt Morgan and his group led to a better definition of a gene. This led to the understanding that different genes controlling different traits are arranged in a linear order on chromosomes. The establishment of this linear order, called linkage maps, in Drosophila maize and other organisms and the discovery of new mutant alleles lent support to the view that the gene is an indivisible
unit of transmission, recombination, mutation and function. The linear order of genes on a chromosome and distances between them deduced from genetic recombination is called linkage or genetic map. The microscopic visualization of chromosomes during the Pachytene stage of meiosis and by the appearance of special chromosomes, the polytene chromosomes, in the salivary glands of larvae of insects like Drosophila supported view that genes were something like ‘beads on a string’. In genetic theory, genes have been considered as

crossover units - hypothetical segments within which crossing over does not occur
breakage units - again hypothetical segments within which chromosome breakage and reattachment do not occur (at any rate not without destruction of one or both fragments)
Mutational and functional units - those minute regions of the chromosomes, which undergo changes to help in development of new alleles.
With a gene being the unit of function, an individual heterozygous for two mutant alleles of the same gene was expected to have only a mutant, but not a wild type phenotype due to the Absence of complementation between alleles. Similarly, heterozygotes for two mutant alleles were not expected to generate a wild type allele by recombination due to the absence of recombination within a gene.

Figure8.55 The wellknown‘onegeneoneenzyme’ hypothesis of BeadleandTatum.
But the genes as indivisible units are not the sole structure of gene, because intra-genic (recombination between two mutant alleles) and inter-genic (when an individual carried two different mutant alleles of the same gene) recombination are possible. In 1948, E B Lewis coined the term Pseudo alleles, to explain the segment of DNA, which takes part in recombination, but fails to produce any phenotype. (Not functional gene copy)
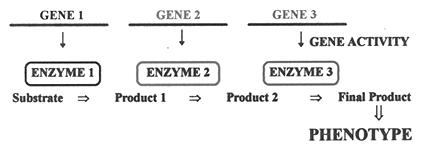
The genes are not mere units with number like appearance, but rather specific chemical and physical entities, which have very complex structure. They are actively involved in functions like their duplication, control of phenotypic characters, mutation and recombination. Seymour Benzer, working at the California Institute of Technology in USA, with his elegant studies on the rII locus of T4 bacteriophage finally established that thegene major properties. Benzer worked in bacteriophages to obtain large numbers of progeny phages and a very convenient selection system to identify even extremely rare phenotypes.
Benzer collected a very large number of mutants (point mutations as well as deletions) at the

This locus displays rapid lysis (causing lysis of more bacterial cells in a given unit of time and therefore, producing larger plaques on a bacterial lawn). The bacteriophage host is the B strain of E.coli
determine if two linkedmutationsaffectingagivenphenotypeareallelic heterozygotes individualsthatcarryboththemutantgenes,( eg.M1and cis(i.e,boththemutantgenes on one chromosomeandboththewildtypealleles(m1+and or in trans(i.e mutantand one thetwomutantsare non affectthesamegene),thetrans On theother theyaffectthesame‘gene’theheterozygote showsamutantphenotype

When we infect the same K12 cell of E. coli with the two rII mutant phages, if the cell was lysed, the two mutants affected different functional units of the rII gene but if the cell was not lysed they belonged to the same functional unit. That means if the mutants could complement each other, then they are different genes.
We call such a functional units within gene as ‘cistron’
Afetusacquiresfingerprints at theage of threemonths
To find out if two different mutants mapping within the rII locus could recombine, we shall allow infection of a B strain cell of E. coli with the two mutant phages: if the two mutants affected different locations within the same gene and if intra-genic recombination could occur between these locations, indicates that such recombination would generate 50% phages that are wild type
Interestingly such recombination is observed and some of the progeny phages were actually, wild type since they successfully infected K12 cells and caused their lyses Thus we can clearly say that a mutational event did not affect a gene as a whole but only a subparts of two defective genes could be recombined to generate a functional wild type allele.

Theseresults with rII mutants thusshowed that with respect to the functionas well as mutation and subdivided. Thus, a genemay include more than one functional unit or cistron.Thislead to the ‘one geneoneenzyme’ hypothesis beingreplaced by the 'onecistron onepolypeptide'concept
For reco and spond almost
Thusgeneis a complex, divisible machine which delivers polypeptides and undergoes rapid and remarkable changes in its workingandindividual components to suite the changingenvironment and

jor role in evolution and sustaining the life on earth.

Figure8.58 Demonstration of intra genicrecombination by S.Benzerin the rII locus T4 bacteriophage. E.coli cells of B strain were simultaneously infected with two phages (rIIm and rIIn) each carrying a different mutation inteh rII gene. Rare recombination between the DNA of the two phages in the interval between the two mutant sites generates a DNA molecule that carries both the mutations and another molecule which is wild type DNA can successfully infects the K12 cells of E.coli
Beardsandafullhead of hairare seen as beingmore aggressiveandlesssociallymature
26.Whymicrobialgenerecombination is called as Horizontalmode of genetransfer?>
27 How wouldyoudifferentiatewhether2bacteriaproducedawildtype by recombination or by mutation?

28.F+donatesplasmid to F-cells, in thelight of evolution,canyouexplainthehelping nature of F+,eventhoughthere is no gain to F+?
29. If at allthegenes to be transferredintoF-cell,howlongtheconjugationshouldcontinue?
30.Doestheconjugationonlytakes up thegoodgenes?

Human Genetics is more complicated than rest other organism genetics
genetic studies are done using Pedigree analysis. It’s the story of human family and along with that inheritance of genes
Pedigree is done to establish the inheritance patters of a gene or a phenotype.
are basically 4 types of inheritance pattern

on the location of genes and their frequency value in gene
Autosomal recessive
Gene is on autosomes
is not passed on to all generations, the phenotype can occur suddenly in child even the parents are normal
Sex linked Dominant : Gene is on X chromosome and is passed to every generation, seen more in females
Sex linked recessive: Gene is on X chromosome and is not passed on to eery generation, it follows the criss cross pattern.
The inheritance pattern is very important to establish the genetic basis of any phenotype.
Pedigree analysis helps us to identify the fashion of inheritance and determination of future genetic conditions in child
By pedigree we have been able to identify the inheritance pattern and thus helping the mankind to protect from possible genetic disorders
Human genetics are not easy to study. Human genome is very complex and we have longer life span, which makes harder to obtain expected 3:1 ratio in inheritance patterns. Thus we use a different tool, called as pedigree analysis, to study inheritance patterns in humans. We have 4 types of inheritance patterns depending on the gene locations.
Table8.5. The table showing the 4 major types of inheritance pattern .
Autosomaldominant
Genes are located on Autosomes
Actdominantin nature
Autosomalrecessive Xlined dominant X-linkedrecessive
Genes are located on Autosomes
Genes located on X chromosomes Genes located on X chromosomes
Act in homozygous recessive Act in dominant Act in homozygous recessivecondition
Ex:AAor Xa Y
In this section, we shall go though each one of these types and let us try to understand the concepts behind them .
We use some kinds of signs to draw
family pedigree charts us to analyse the data very easily and efficiently.
you start your analysis is called Proband. ( Propositus) He first comes to your attention. Using his condition as reference you start drawing familydisease patterns blood relatives is ous marriage
There are basically 4 types of major inheritance pattern in humans. One depends on the presence of the gene on Autosomal chromosomes or on sex chromosomes. Other one is the inheritance pattern, whether the gene is getting inherited in dominant manner or in recessive nature.
The inheritance of a gene does not follows same pattern always. The genetic factors such as Penetrance, expressivity, co-dominance, epistasis, Pleotropy, Imprinting, Environmental effects influence the inheritance. Thus they are largely ignored in classical pedigree
analysis.
Apart from there, Y chromosomal inheritance and extra chromosomal inheritance are also observed. But there has been no solid proof for Y linked inheritance of any gene, in fact the maleness is itself is Y linked. Hence whenever we say sex linked inheritance, it means X chromosome inheritance

As, we already learnt, these alleles act in dominant condition. If so, all the people in a family should get the disease, then why it is not the case? Because, the concept of dominance and recessive is only due to its expression patterns. An allele is said to be dominant, if it acts in Dominant condition, and not if it is found in most cases.
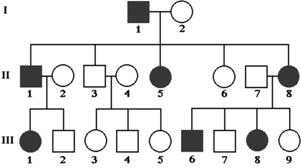
Disease tends to appear in every generation.

Affected parents transmit the phenotype to both sons and daughters.

It occurs in both sexes in equal proportions.
Abnormal child will have affected parents.
Most affected individuals are heterozygotes.

In offspring born to one affected parent and one normal parent, show 1:1 abnormal to normal ratio.
1. Pseudochondroplasia,
People with genotype ‘dd’ are normal and people with genotype ‘Dd’ and ‘DD’ are dwarf.
The allele B interferes with bone growth.
Interestingly, the gene acts in dominant condition, but still the disease is rare in the population.
It is a rare dominant disorder , which causes extra fingers in hand or toes in legs
It is caused by the allele P.
The expression pattern of P allele is not uniform
In some cases it causes individuals to have 6 fingers and some case it causes 7 fingers.
It is a rare dominant disorder caused by mutation in c-kit gene
It appears as white patches on the body. It is different form albinism in molecular mechanism.
It is caused due to improper movement of melanocytes from dorsal to ventral surface during development
Because of amutationand in-breeding,thetown of Sao Pedro,Brazilhasa 10%rate of twinkids
The phenotype is caused by recessive condition of an allele, ‘aa Whenever a person gets 2 copies of recessive gene, he will show the character. Autosomal Recessive is very common and very few people in population will show the disease. The person with heterozygous condition will be called a Carrier for the disease, as he does not show the disease, but can pass on the affected gene to next generation. Important characteristics:
The disease appears in the progeny of unaffected parents
It occurs in both male and female progeny in equal proportion
It may skip generations
The probability of occurrence in progeny of unaffected parents in ¼

.Autosomalrecessive condition.Nottheskipping of generationandverylowfrequency
Most common disorders: Phenylketonuria (PKU), Albinism, Cystic fibrosis
These genes are located on the X chromosomes
They act when present in dominant state.
They are found more in females than in males.
The disease is never passed from father to son.
Affected males pass the condition to all their daughters, but no son gets the disease.
Affected heterozygous females and normal male pass the disease to half sons and daughter.
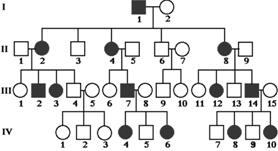
Most common disorder includes: Hypophosphatemia, a type of vitamin-D resistant rickets.
It occurs more to females than males
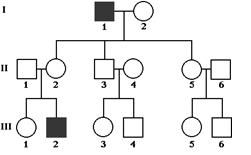
Disease occurs when found in homozygous recessive condition.
Xc in females and X
recessive condition is very rare, then only males will be affected.
of an affected male not be affected. But all his daughters are carriers.
bear a recessive allele, which is passed on to next generation.
Disease is never passed from affected father to son.
Occurs more to males, almost exclusively
Red-green color blindness.
People cannot distinguish red from green.
.SexlinkedDominant condition.Nothighfrequency of femalesthanmales
.Sexlinkedrecessive condition.Nottheskipping of generationandparticularlyfound in males, in morefrequency
The genes coding for the cone cells, which recognize red and green color is located on X chromosomes.
It affects more in males, than females
acts in recessive condition.
2. Hemophilia
Failure of blood to clot.
Caused by absence / malfunction of factor VIII.
famous case is in royal families in Europe.

starts at age of 6, patient has
depend on wheelchair by age 12 and death by 20.
improper/ absence of gene encoding muscle protein
Androgen insensitivity syndrome.
Individuals possess 44+XY condition,
Dystrophin.
still show female characters.
show female genitalia and non-developed uterus and vegina
Mutation in androgen receptor
Solved Problem : 1
Occurs in most cases and introduced by male from other family and passing on to every generation
Either X linked or Autosomal dominant
Affected father, is not passing on to any of his sons
Ans: It is X-linked dominant.
When a disease allele is known to be present in a family, knowledge of simple gene transmission patterns can be used to calculate the probability of prospective parents’ having a child with the disorder. For example, a married couple finds out that each had an uncle with Tay-Sachs disease (a severe autosomal recessive disease). The pedigree is as follows:
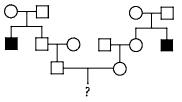
The probability of their having a child with Tay-Sachs can be calculated in the following way. The question is whether or not the man and woman are heterozygotes (it is clear that they do not have the disease) because if they are both heterozygotes then they stand a chance of having an affected child.
1.The man’s grandparents must have both been heterozygotes T/t because they produced a t/t child (the uncle). Therefore, they effectively constituted a monohybrid cross. The man’s father could be T/T or T/t, but we know that the relative probabilities of these genotypes must be 1/4 and 1/2, respectively (the expected progeny ratio in a monohybrid cross is 1/4 T/T, 1/2 T/t, and 1/4 t/t). Therefore, there is a 2/3 probability that the father is a heterozygote [calculated as 1/2 divided by ( + 1/4+1/2)]. 2.The man’s mother must be assumed to be T/T, since she married into the family and disease alleles generally are rare. Thus if the father is T/t, then the mating to the mother was a cross T/t × T/T and the expected progeny proportions are 1/2 T/T and 1/2 T/t.
3.The overall probability of the man’s being a heterozygote must be calculated using a statistical rule called the product rule, which states that the probability of two independent events both occurring is the product of their individual probabilities. Hence the probability of the man’s being a heterozygote is the probability of his father’s being a heterozygote times the probability of the father having a heterozygous son, which is 2/3 × 1/2 = 1/3.
4. Likewise the probability of the man’s wife being heterozygous is also 1/3.
they are both heterozygous (T/t),
the probability of their having a t/t child is 1/4, so overall the probability of the couple having an affected child is 1/3 × 1/3 × 1/4 = 1/36; in other words, a 1 in 36 chance.
Solved Problem : 2
PKU is a human hereditary disorder resulting from inability of body to process phenylalanine. A couple intends to have children but consults a genetic counselor because the man has a sister with PKU and the woman has a brother with PKU. There are no other known cases in their families. They ask genetic counselor to determine the probability that their first child will have PKU. What is this probability?
What can we deduce?
Let us take p P normal allele.
Then man’s sister and woman’s brother will have “ ” genotype ( because they are affected) The man and woman should be heterozygous to have a child with PKU. The grandparents are normal and carriers. Then what's the chance that the man and woman are heterozygous? Thus they can have 3 possible combinations, PP, In these only 2 are heterozygous, thus probability of man being heterozygous is 2/3.
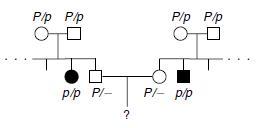
Probability of man and woman to be heterozygous = 2/3 x 2/3 = 4/9
Probability of heterozygous parents to have affected child = ¼
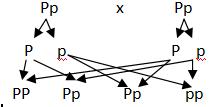
Thus the probability that the parents are heterozygous and first child to be affected is = 4/9 x ¼ = 1/9

Ans: 1/9
Humans hav and draw human genetic in humans, it is not possible to se difficulties, LOD score ha
Whenever together in an organism, in can be by due to 2 factors: 1. Chance 2. The genes might be linked. We need to find what exactly has happened, so that we can determine the genetic map and human genome sequencing. Let us see how this is done, with the help of LOD, a statistical tool. It calculates the 2 different probabilities for a given trait.

First probability is calculated, with no linkage; only independent assortment.
Second probability, with linkage.
Using formula, the LOD score is calculated.
NR - The number of non-recombinant offspring
R - The number of recombinant offspring.
Unit 8
0.5 is used in the denominator is that any alleles that are completely unlinked have a 50% chance of recombination, due to independent assortment.
Theta is the recombinant fraction, it is equal to R / (NR + R)
Interpreting LOD score
•LOD score >3.0 = linkage.
•LOD score = +3 indicates did not occur by chance. i.e, Linked.
•LOD score less than -2.0 exclude Linkage. i.e, chance
The following pedigree will be used to demonstrate another method developed to determine the distance between genes. This approach has been widely adapted to various system and genetic programs have been developed based on this technique. First let's look at the pedigree in figure 8.64A
Several points can be drawn from this pedigree. Even though we are working with the same two genes, nail-patella and blood type, in this pedigree the dominant allele seems to be coupled with the A blood type allele. Remember in the previous example, the dominant nail-patella allele was linked with the B allele. This is an important point in genetics --- not all linkages between alleles of two genes are found to be constant throughout a species. Why??? Because at some point in the lineage of this family, the disease (nail-patella) allele recombined and became linked to a different blood type allele. In even other lineages, the nail-patella causing allele is linked to the O blood type allele.
linkage in
Next let's determine the linkage distance between the two genes. As you can see, we have one recombinant among the eight progeny. This gives us a recombination frequency of 0.125 and a distance of 12.5 cM.
We will now introduce a new method to calculate linkage distances called the Lod Score Method. The method developed by Newton E. Morton is an iterative approach were a series of lod scores are calculated from a number of proposed linkage distance. Here is how the method works. A linkage distance is estimated, and given that estimate, the probablity of a given birth sequence is calculated. That value is then divided by the probability of a given birth sequence assuming that the genes are unlinked. The log of this value is calculated, and that value is the lod score for this linkage distance estimate. The same process is repeated with another linkage distance estimate. A series of these lod scores are obtained using different linkage distances, and the linkage distance giving the highest Lod score is considered the estimate of the linkage distance. The following is the formula for the lod score:
The above example will be used to demonstrate the principle. We will first use 0.125 as our estimate of the recombination fraction. In this first birth sequence, we have an individual with a parental genotype.The probablity of this event is (1 - 0.125). Because there are two parental types, this value is divided by two to give a value of 0.4375. In this pedigree we have a total of seven parental types. We also have one recombinant type. The probability of this event is 0.125 which is divided by two because two recombinant types exist. What would the sequence of births be if these genes were unlinked? When two genes are unlinked the recombination frequency is 0.5. Therefore, the probability of any given genotype would be 0.25.

Now let's put the whole method together. The probability of a given birth sequence is the product of each of the independent events. So the probability of the birth sequence based on our estimate of 0.125 as the recombination frequency would be equal to (0.4375)7(0.0625)1 = 0.0001917. The probability of the birth sequence based on no linkage would be (0.25)8 = 0.0000153. Now divide the linkage probability by the non-linkage probability and you get a value of 12.566. Next take the log of this value, and you obtain a value of 1.099. This value is the lod score.
We can calculate this relative likelihood under the assumption that the two genes are tightly linked with no recombination occurring between them at all, i.e. with the recombination fraction θ = 0. Or we can calculate the relative likelihood for any other value of θ that we care to choose. The value reaches a maximum positive value is then our best estimate of the distance between the two loci. The value of Zmax is a measure of our confidence in the result. If Zmax > 3 then we conventionally take the linkage to have been proved. Conversely, if Z <we take the linkage to have been disproved for that value of θ
The data are often displayed in the form of a table of Z calculated for a range of theta from 0 to 0.5. e.g.


Notice in this example that Z has sunk to minus infinity at θ = 0. This will be the case if there has been at least one recombination event between the genes, they cannot then be zero distance apart. It is often helpful to display the data graphically as shown in the red curve on the right. The maximum value of Z occurs at a recombination fraction of 0.1 and it is greater than 3. Therefore we can feel confident that the two genes are indeed linked and our best estimate for the genetic distance between them is θ = 0.1 (equivalent t0 0cM). Compare this with the blue curve plotted for two unlinked genes. In this case Z is always negative and the best estimate of θ is 0.5 i.e. unlinked.

Genome is organized into several standard chromosomes, each with specific characters and structures. A Karyotypes is an organized profile of a person's chromosomes. In a Karyotypes, chromosomes are arranged and numbered by size, from largest to smallest. This arrangement helps scientists quickly identify chromosomal alterations that may result in a genetic disorder. Karyology (the study of whole set of chromosome) describe the number of chromosomes, and what they look like under a light microscope. Attention is paid to their length, the position of the centromere,
Figure8.65 A. The types of chromosomesbased on thelocation of centromere.Notethechange in shape.
Unit 8
banding pattern, any differences between the sex chromosomes, and any other physical characteristics. The preparation and study of Karyotypes is part of cytogenetic The chromosomes are rearranging in a standard format known as a karyogram or ideogram.
Preparing a Karyotype:
Metaphase cells are required to prepare a standard Karyotypes, and virtually any population of dividing cells could be used. Blood is easily the most frequently sampled tissue,
A sample of blood is drawn and coagulation prevented by addition of heparin.

Mononuclear cells are purified from the blood by centrifugation through a dense medium that allows red cells and granulocytes to pellet, but retards the mononuclear cells (lymphocytes and monocytes).
The mononuclear cells are cultured for 3-4 days in the presence of a mitogen like phytohemagglutinin, which stimulates the lymphocytes to proliferate rapidly.

At the end of the culture period, when there is a large population of dividing cells, the culture is treated with a drug such as colcemid, which disrupts mitotic spindles and prevents completion of mitosis. This greatly enriches the population of metaphase cells.
The lymphocytes are harvested and treated briefly with a hypotonic solution. This makes the nuclei swell osmotically and greatly aids in getting preparations in which the chromosomes don't lie on top of one another.
The swollen cells are fixed, dropped onto a microscope slide and dried.
Slides are stained after treatment to induce a banding pattern. Once stained slides are prepared, they are scanned to identify "good" chromosome spreads (i.e. the chromosomes are not too long or too compact and are not overlapping), which are photographed.
Spectral Karyotypes (SKY technique)
Spectral karyotyping is a molecular cytogenetic technique used to simultaneously visualize all the pairs of chromosomes in an organism in different colors.
Fluorescently-labeled probes for each chromosome are made by labeling chromosome-specific DNA with different fluorophores.
there are a limited number of spectrallydistinct fluorophores, a combinatorial labeling method is used to generate many different colors.
Spectral differences generated by combinatorial labeling are captured and analyzed by using an interferometer attached to a fluorescence microscope.
Image processing software then assigns a pseudo color to each spectrally different combination, allowing the visualization of the individually colored chromosomes.
To study chromosomal aberrations
To study Cellular function
Figure8.65 B. The HumanKaryotyping. The 46 chromosomeshavebeenarrangedaccording to their shapeandsize. We can see 7groups of chromosomes according to theirsizes.Chromosome1 is largestoneand chromosome 22 is thesmallest.XandYchromosomesare arranged at theend.
chromosome
a woman from becoming pregnant or causing
types of
Ever increasing human genetic disease is a greatest challenge to human population. The no. of diseases with genetic basis are difficult to diagnose and difficult to cure. The search for cause of these genetic diseases at biochemical level has been an active area of research in biology. In this chapter, let us analyze some of the
Initial studies were done by British biologist, Archilbald Garrod. He worked on Alkaptonuria, and he found out that it is a genetic disorder arising from metabolic errors. Thus the initial scientists had correlated that certain enzyme defect caused by gene mutations, can cause genetic disorders. A gene usually codes for one or more enzymes. Enzymes carry out some important functions and catalyze some important biochemical reactions. Any defect in genetic level, has direct effect on biochemical reactions. These result in genetic defects and inherited disorders. The failure of an enzyme in the metabolic sequence to perform its function results in the accumulation of the un-reacted metabolites. Such blocks in metabolic pathways results in serious problems in cellular level and problems to the individual organism.
Now, lets us study the important molecular aspects of genetic disorders: in the same gene can lead to severe phenotypes. Any protein or regulatory enzyme can be altered. Regulatory mutations, it can be a point mutations occurring in reading frame of genes which are regulatory in function Mutations affecting the protein product , point mutations in some important genes which will alter the protein structure and function , deletion mutations of some gene, which induce complete absence o , the tri nucleotides occur more and more in genome, causing problems in gene regulation, e.g: neuropsychiatric disorders
1. Charcot-Marie-Tooth disease.
Some mRNAs recruit ribosomes directly to initiating AUG codons through 5’ un-translated region
Mutation of the IRES of the c-myc gene causes increased translation of this oncogene, leading
2. Hyperferritinemia / cataract syndrome (HHCS)

caused by mutations that affect an iron-responsive element (IRE) in the 5’ UTR of the L-ferritin
Mutation of this IRE element in the 5’ UTR results in abnormally high L-ferritin production
3. Hereditary thrombophilia (increased tendency for the blood to clot)
It is caused by a single nucleotide substitution in the 3’ UTR,
This mutation leads to excess production of thrombin mRNA and protein
Pulmonary embolism is the most common cause, in the industrialized world, of maternal death during pregnancy or in the period following delivery.
About 70% of women who present with venous thrombo embolism during pregnancy are carriers of hereditary or acquired thrombophilia
medical condition
following characteristics.
Due to stepwise expansion of unstable triple repeats
first step is the formation of a
that has a normal phenotype but is unstable
Premutation then expands in a subsequent generation to a much greater length and further instability
anticipation is a hallmark of trinucleotide repeat
Autosomal dominant disease
by myotonia and progressive muscle weakness
most common form of adult-onset muscular dystrophy

cardiovascular, and ocular (cataracts)
with cognitive changes, including mental retardation
by mutations in the DM-1 or myotonin protein kinase gene which is expressed in brain, heart, and muscle
gene has a 3’-UTR CTG repeat that ranges from 5-30 repeats
have expanded numbers of repeats, up to many hundreds, that lead to decreased mRNA levels
2. Fragile X syndrome (FRAXA)
X-linked Dominant
in one of 1 ,250 male births and is the second most common cause of mental retardation
Associated with moderate to severe mental retardation
Disease shows anticipation , increasing “penetrance” in succeeding generations
Passage through female can increase risk to next generation
Females with one affected chromosome and males with premutations can show mild cognitive defects and schizotypal symptoms
Caused by mutations in the FMR-1 gene which is expressed in brain and testes at highest levels, and widely in the embryo
Mormal gene has a 5’-UTR CGG repeat ranges from 6-55 repeats
Patients have expanded numbers of repeats, up to thousands results in transcriptional silencing
3. Spinal and bulbar muscular atrophy (Kennedy’s)
X-linked; rare
late onset form of motor neuron degeneration associated with mental retardation and insensitivity to androgens
caused by mutations in the androgen receptor gene
normal gene has a CAG repeat that is polymorphic in the population and that ranges from 13-28 repeats
patients have expanded numbers of repeats, up to 39-60 repeats
CAG repeat is translated into a polyglutamine tract in the protein
disease shows anticipation, but the severity of the illness does not correlate with the degree of expansion
4. Huntington’s disease
Autosomal dominant disease
juvenile to late adult onset
associated with involuntary movements (chorea), behavioral disturbances, and cognitive impairment
caused by mutations in the 1T15 gene (whose function is unknown)
normal gene has a CAG repeat ranges from 11-35 repeats
Patients have expanded numbers of repeats (>35 repeats)
CAG repeat is translated into a polyglutamine tract in the protein
The disease shows anticipation
Unit 8
1.Spinocerebellarataxia type I (SCAI) (CAG polyglutamine)
2.Dentatorubral-pallidoluysian (DRPLA) (CAG polyglutamine)
3.Machado-Joseph’s disease (MJD/SCA3) (CAG polyglutamine)
4.FRAXE mental retardation (GCC)
5.Jacobson’s disease (GCC)
1.Glycogen storage disease
Absence of Glucose-6-phosphatase.
This enzyme produces blood sugar from glycogen
Patients show severe cases of hypoglycemia
Symptoms include
If the persons lives until adult hood, he will show gouty arthritis.
These patients have lacticacidemia and ketonemia.
in renal retention of uric acid and increased Purine biosynthesis.
2. Alcaptonuria
Also called as black urine syndrome.
Lack of an enzyme
This enzyme is involved in Tyrosine amino acid metabolism
Absence of this enzyme, accumulates the Homogentisic acid, which is excreted in urine.
3.Colorectal cancer
One of the most common cancers and second cause of death in developed countries.
In addition to environmental effects, various genetic predeposition plays significant role in its development.
Deletion of APC gene is most common in the patients.
Improper splicing of this gene causes multiple adenomatous polyposis phenotypes.
Microsatellite instability also increases occurrence of disease.
4.Male infertility
Y-chromosomes are associated with male infertility, as males contain Y chromosome specifically
Micro deletion in the 3 non-overlapping regions of the Y-chromosome is responsible for male infertility
The deletion size can
People with azoospermia and severe oligozoospermia sow these deletions.
Normal people show no deletion.

1.Shape of face (probably polygenic)
2.Cleft in chin
3.Hair curl (probably polygenic) Assume incomplete dominance
4.Hairline
5.Eyebrow size
6.Eyebrow shape
7.Eyelash length
8.Dimples
9.Earlobes
10.Eye shape
11.Freckles
12.Tongue rolling
13.Tongue folding
14.Finger mid
Oval dominant, square recessive
No cleft dominant, cleft recessive
Curly: homozygous Wavy: heterozygous Straight: homozygous
Widow peak dominant, straight hairline recessive
15.Hitch hiker thumb
16.Bent little finger
17.Interlaced fingers
18.Hair on back of hand
19.Tendons of Palmar Muscle
Left thumb over right dominant, right over
Hair dominant, no hair recessive
Two tendons dominant, three tendons recessive
Human Phenotypes and their Inheritance pattern

Recombinant DNA technology has immensely contributed to the diagnosis of genetic diseases in fetuses
is also used in the treatment of some genetic disorders in animals.
The basic approach to diagnosis of disease in fetus is to obtain fetal cells by amniocentesis (that is, a sample of amniotic fluid taken by inserting a needle into the amniotic cavity)
test for biochemical defects.
genetic defects may not be recognizable in the
amniotic fluid cells obtained by amniocentesis through
routine biochemical methods only
the case of lethal recessive mutations, gene cloning provides the potential for direct diagnosis.
After a normal gene has been cloned, the nucleotide sequence and restriction endonuclease map will be determined
Then it can be tested detect the mutations responsible for that particular defects.
The normal gene can be used as a probe to isolate or locate the corresponding gene from the DNA of the fetal cells
The restriction fragment length polymorphism (RFLP)

The restriction fragment length polymorphism (RFLP) is now being used as a tool in clinical diagnosis of human and animal diseases.
This involves prenatal diagnosis using amniotic fluid, chorionic villus, northern and southern blotting, electrophoresis and hybridization.
The use of RFLP in the diagnosis of diseases reduced the stress of gene disorders
By analyzing the RFLP patterns we can predict the occurrence of disease in new born.
Genetic engineering techniques are the most sensitive and reliable human DNA fingerprinting for use in forensic analysis.
is the process of the selection for a particular trait using genetic markers.
MAS selects the genes with increased accuracy
MAS helps in identification of carriers of genes for resistance.
example is the gene responsible for trypanosoma tolerance in cattle.
Critical thinking Questions
Is
If picture?
thegeneticphenomenon large. Canyounameafew of them?
How do youthinkthepedigree of Co-dominantalleleswouldbehave?
we findoutwiththehelp of pedigreelocation of genes on chromosomes?


Genetics is a science of variation. Without variation, there is no evolution, there is no survival. A flower’s petal can be red or white, a bird’s wing can be short or long, we can be tall or dwarf. In other terms we can have all kinds of variations some are very distinct (Tall / dwarf; Red/ white) or they can be continuous (red, pink, light red, pinkish white…). Thus genetic variations can be continuous or discontinuous. Quantitative genetics is the branch of genetics, which deals with all these variations in statistical way.
Just like snowflakes, no two fingerprints are alike, even those of identical twins. Yet, research shows that the patterns of ridges on our fingers and palms are inherited. What factors would produce such a variety of phenotypes? Is the trait encoded by more than one locus? Are there environmental factors involved? Is there a relationship between a person's fingerprints and another trait, such as hair color or blood type? In this chapter, you have learned the answer to questions such as these. In the activity, you can apply what you've learned as you investigate whether a relationship exists between fingerprint patterns and high-blood pressure.
In natural populations, variation in most characters takes the form of a continuous phenotypic range rather than discrete phenotypic classes. In other words, the variation is quantitative, not qualitative. Mendelian genetic analysis is extremely difficult to apply to such continuous phenotypic distributions, so statistical techniques are employed instead. A major task of quantitative genetics is to determine the ways in which genes interact with the environment to contribute to the formation of a given quantitative trait distribution.
The genetic variation underlying a con-

tinuous character distribution can be the result of segregation at a single genetic locus or at numerous interacting loci that produce cumulative effects on the phenotype. Thus quantitative genetics depends on the no. of genes and no. of loci, involved in the character development.
There are 2 major types of characters: 1. Qualitative and 2. Quantitative traits. Qualitative, or discontinuous, characteristics possess only few distinct phenotypes. These characteristics are the types studied by Mendel and have been the focus of our attention thus far. However, many characteristics vary continuously along a scale of measurement with many overlapping phenotypes. They are referred to as continuous characteristics; they are also called quantitative characteristics because any individ-
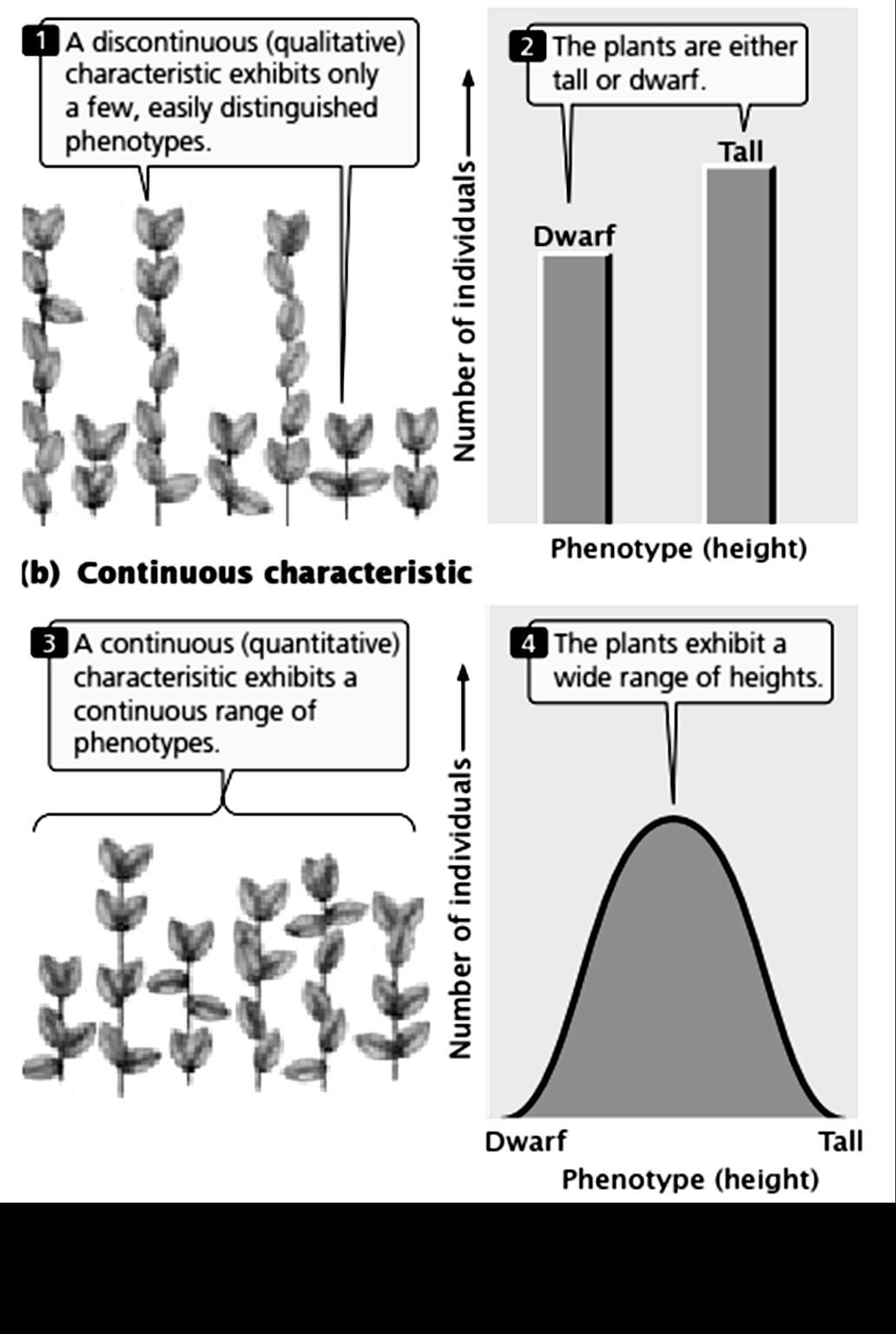
Unit 8
ual’s phenotype must be described with a quantitative measurement. Quantitative characteristics might include height, weight, and blood pressure in humans, growth rate in mice, seed weight in plants, and milk production in cattle.
In general, the distribution of quantitative traits values in a population follows the normal distribution. These curves are characterized by the mean (mid-point) and by the variance. There are 3 types of continuous characters.
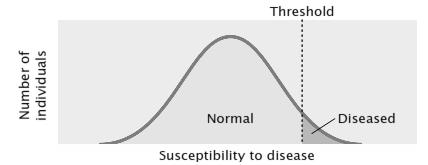
1. Continuous trait: can take on any value: height, for example.
2. Meristic trait: Can take on integer values only: number of bristles, for example.
3. Threshold trait: Has an underlying quantitative distribution, but the trait only appears only if a threshold is crossed.

Sometimes, we observe that a phenotype is coded by multiple genes, thus our old concept of one gene, one enzyme does not valid. This is called as Polygenic (Poly = Many, Genic = genes) inheritance. The inheritance patterns observed by polygenic inheritance will be slightly different, than the Mendelian. Here, the no. of genes which code for the phenotype matters much.
This type of hibit continuous, quantitative, or metrical variation. heritance pattern is polygenic or quantitative.
The system is termed an additive model because each allele adds a certain amount to the phenotype.

we can predict the distribution of genotypes and phenotypes expected from
model with linked loci segregating two alleles each Thus the character depends on the no. of loci participated in character development.
When a strain of heavy mice was crossed with a lighter strain, the F1 were of intermediate weight. When these F1 were interbred, a continuous distribution of adult weights appeared in the F generation. Since only about one mouse in 250 was as heavy as the heavy parent stock, we could guess that if an additive model holds, then four loci are segregating. This is because we expect 1/(4)n to be as extreme as either parent
Figure8.68 The thresholdcharactershowsonly2 possibilities,eitherPresent or Absent, but theyhavea thresholdlevel.
Unit 8
One in 250 is roughly 1 / (4)4, = 1/256. So out of 256 plants, only 1 plant will be just like one of the parent. Rest all 255 will be having all other variations from the main phenotype.
To illustrate how multiple genes acting on a characteristic can produce a continuous range of phenotypes, let us examine one of the first demonstrations of polygenic in-

heritance.
Nilsson-Ehle studied kernel color in wheat and found that the intensity of red pigmentation was determined by three unlinked loci, each of which had two alleles. Nilsson-Ehle obtained several homozygous varieties of wheat that differed in color.
Like Mendel, he performed crosses between these homozygous varieties and studied the ratios of phenotypes in the progeny. In one experiment, he crossed a variety of wheat that possessed white kernels with a variety that possessed purple (very dark red) kernels and obtainedthe following results: Nilsson-Ehle interpreted this phenotypic ratio as the re two loci. He proposed that there were two alleles at each locus: one that produced red pigment and another that produced no pigment.
We’ll designate the alleles that encoded pigment A- and B- and the alleles that encoded no pigment A and BNilsson-Ehle recognized that the effects of the genes were additive. Each gene seemed to contribute equally to color; so the overall phenotype could be determined by adding the effects of all the genes, as shown in this table.
Notice that the purple and white phenotypes are each encoded by a single genotype, but other phenotypes may result from several different genotypes.
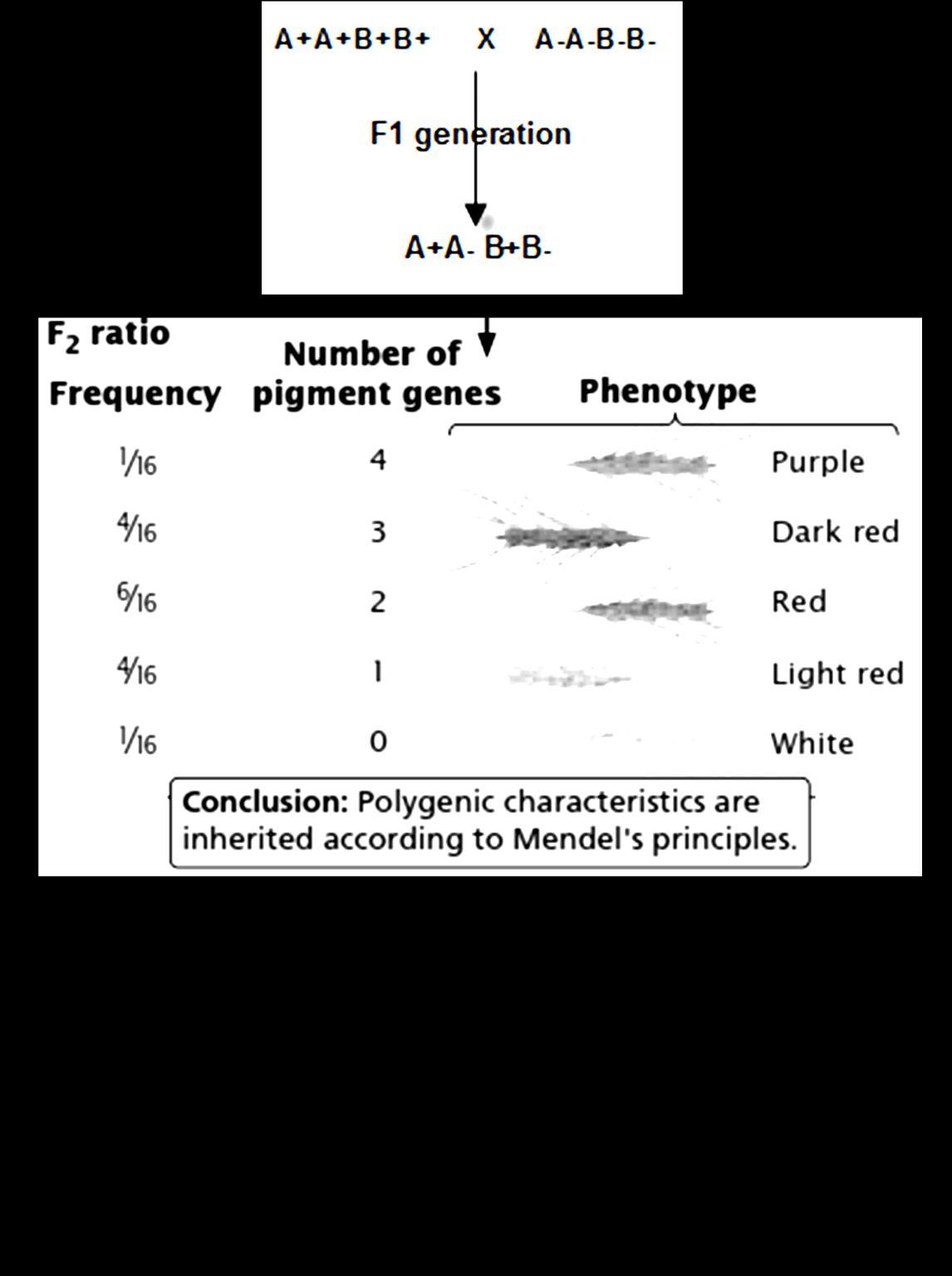
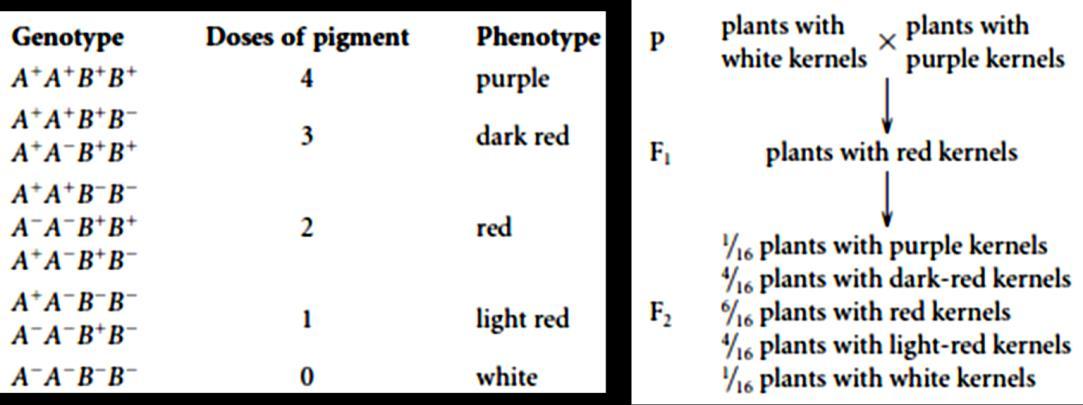
From these results, we see that five phenotypes are possible when alleles at two loci influence the phenotype and the effects of the genes are additive.
When alleles at more than two loci influence the phenotype, more phenotypes are possible, and this would make the color appear to vary continuously between white and purple. If environmental factors
influenced the characteristic,
the same genotype would vary somewhat in color, making it even more difficult to distinguish between discrete phenotypic classes.
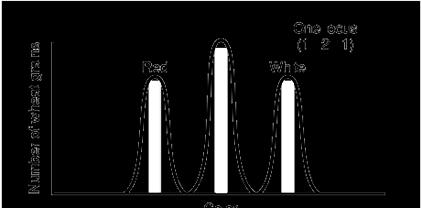
color in Nilsson-Ehle’s crosses, and only a few loci encoded color; so Nilsson-Ehle
environment played little role in determining
able to distinguish among the different phenotypic classes. This ability allowed him
see the Mendelian
of the characteristic.
Let’s now see how Mendel’s principles explain the ratio obtained by Nilsson-Ehle in his F progeny. Remember that NilssonEhle crossed a homozygous purple variety with homozygous white variety.
The number of loci, involved in genetic inheritance is most important in determining the characters and its variability. If only one gene loci is involved, we often find possible conditions, one with all homozygous dominant state, one with all homozygous recessive state and the last one with heterozygous dominant state. The ratio of the phenotype = 1:2:1 (Figure 8.71) 2 locus control: Here genes at 2 different locations (in simple notion, 2 genes). They act in many different ways to produce 5 phenotypic in ratio 1:4:6:4:1. (Figure 8.72)
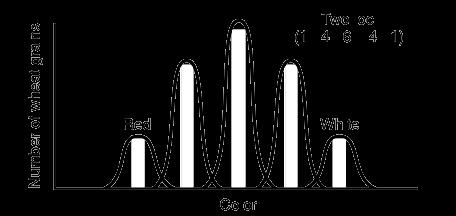
Plant and animal breeders want to improve the yields of their crops to the greatest degree they can. They must choose the parents of the next generation on the basis of this generation’s yields; thus, they are continually performing selection experiments. Breeders have two economic problems. They cannot pick only the very best to be the next generation’s parents because (1)they cannot afford size of a crop by using only very few select parents and (2) they must avoid inbreeding depression, which occurs when plants are self-fertilized or animals are bred with close relatives for many generations. After frequent inbreeding, too much homozygosity occurs, and many genes that are slightly or partially deleterious begin to show themselves, depressing vigor and yield
Breeders often calculate a heritability estimate, a value that predicts to what extent their selection will be successful. Heritability is defined in the following
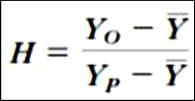
heritability

Y
Y
From this equation, we can see that heritability is the gain in yield divided by the amount of selection practiced
, Y0 - Y, is the improvement over the population average due to YP -Y which is the amount of difference between the parents and the population average. If there is no gain, Y0 = Y then the heritability will be zero, and breeders will know that no matter how much selection they practice, they will not improve their crops and might as well not waste their time. Since this value is calculated after the breeding has been done, it is referred to as realized heritability


This heritability is the proportion of the total phenotypic variance caused by additive genetic effects. The heritability of most interest to plant and animal breeders because it predicts the magnitude of the response
Under selection. Heritability depends on both Environment and genotype of parents and expected correlations between relatives as
In which robs is the expected correlation. The expected correlation In human beings, finger-ridge counts (fingerprints), have a very high heritability; there seems to be very little environmental interference in the embryonic development of the ridges .Monozygotic twins are from the same egg, which divides into two embryos at a very early stage. They have identical genotypes. Dizygotic twins result from the simultaneous fertilization of two eggs. They have the same genetic relationship as siblings. (However, environmental influence may different; they may be treated differently by relatives and friends.) The data therefore suggest that human finger ridges are almost completely controlled by additive genes with a dominance variation. Few human traits are controlled this simply.

Skin color is a quantitative human trait for which a simple analysis can be done on naturally occurring mating. Certain groups of people have black skin; other groups do not. Many of these groups breed true in the sense that skin colors stay the same generation after generation within a group; when groups inte marry and produce offspring, the F are intermediate in skin color. In turn, when F 1 individuals intermarry and produce offspring, the skin color of the F is, on the average, about the same as the F2 but with more variation. The data are consistent with a model of four loci, each segregating two alleles. At each locus, one allele adds a measure of color, whereas the other adds none. We are expecting 4 loci in human skin color.
In human beings, twin studies have been helpful in estimating the heritability of quantitative traits. One way of looking at quantitative traits is by the concordance among twins. Concordance means that if one twin has the trait, the other does also. Discordance means one has the trait and the other does not. Table 18.8 shows some concordance values. High concordance of monozygotic as compared with dizygotic twins is another indicator of a trait. Concordance values for measles susceptibility and handedness, which are similar for both Monozygotic and dizygotic twins, demonstrate the environmental influence on some traits. Some
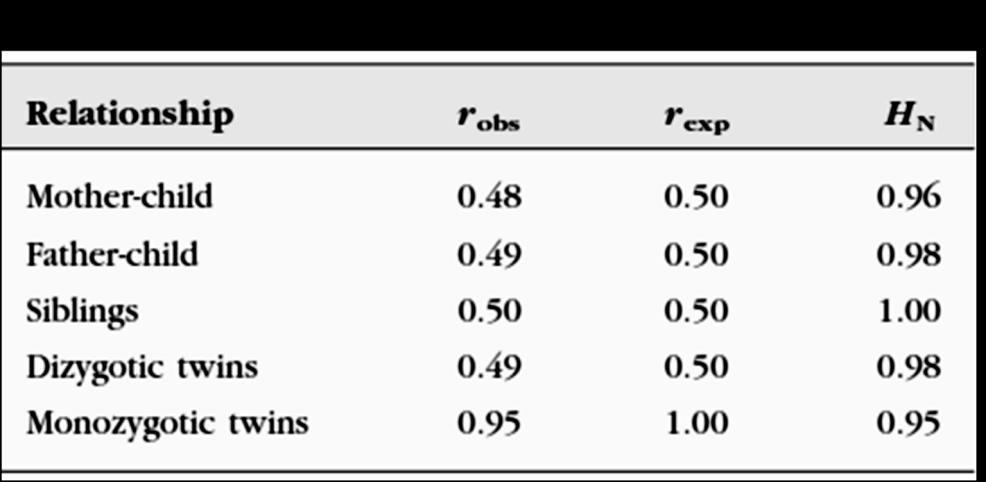
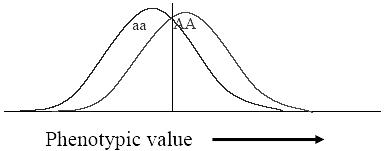
monozygotic twins (MZ) have been reared apart. The same is true for dizygotic twins (DZ) and non-twin siblings. IQ (intelligence quotient) is a measure of intelligence highly correlated among relatives, indicating a strong genetic component. In three studies of monozygotic twins reared apart, the average correlation in IQ was 0.72. In thirty-four studies of monozygotic twins reared together, the average correlation in IQ was 0.86; dizygotic twins reared together have an average correlation of 0.60 in IQ. Thus, it is clear that there is a genetic influence IQ. However, experts disagree strongly on the environmental role in shaping IQ and the exact meaning of IQ as a functional measure of intelligence
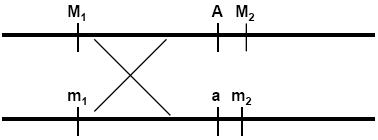

Mapping the location of a standard locus is conceptually relatively easy. We look for associations of segregate wit tance between loci by the However, with
mapping because genes contributing to the phenotype are often located continuous phenotype will be controlled by loci linked to numerous other loci, many unlinked to each other. However, with the advent of molecular techniques, it has become feasible to map polygenes We look at a population of organisms and note various RFLPs or other molecular markers. We then look for the association of a marker and a quantitative trait. If an association exists, we can gain confidence that one
or more of the polygenes controlling the trait is located in the chromosomal region near the marker. The closer the polygenes are to the markers, the more reliable our estimates are, because they depend on few crossovers taking place in that population. With many crossovers, the association between a particular marker and a particular effect diminishes. Since we don’t know immediately from this method whether the region of interest has one or more polygenic loci, a new term has been coined to indicate that ambiguity. Instead of talking about polygenic loci directly, we talk of quantitative trait loci.
QTL = a gene or chromosomal region that affects a quantitative trait. It must be polymorphic (have allelic variation) to have an effect in a population and it must be linked to a polymorphic marker allele to be detected
QTL: underlying genes controlling quantitative traits, measured with large error effects. The result is continuous phenotypic distributions
The QTL effect:
The additive effect of a QTL allele = a Average value of random lines from a cross between AA and aa parents = P
Mean of AA lines is P + a
Mean of aa lines is P – a Effect of a marker linked to a QTL: Recombination between M and A is R n RILs derived from MmAa F1, individuals with MM marker genotype are made up of 2 QTL genotypes: AA & aa If M and A are tightly linked, most are AA. If M and A are far apart, as many as half are aa, So, the effect of marker M is a function of:
distance from the QTL
size of the QTL effect
QTL mapping with molecular markers: DNA markers used to map useful genes using recombination frequencies of linked genes: co-segregate with them tightly linked to QTL detected by ANOVA
QTL mapping strategies: All marker-based mapping experiments have same basic strategy:
1. Select parents that differ for a trait
2. Screen the two parents for polymorphic marker loci
3. Generate recombinant inbred lines (can use F2-derived lines)
4. Phenotype (screen in field)
5. Contrast the mean of the MM and mm lines at every marker locus
6. Declare QTL where (MM-mm) is greatest How is QTL mapping best used?

QTL mapping = very inaccurate for detecting, localizing, and estimating the effect size of genes with a small effect
If repeatability QTL phenotyping experiment = low QTL map very unreliable
Unit 8
QTL mapping works very well to find single genes with large effects
QTL mapping requires a phenotypic screening system with high H

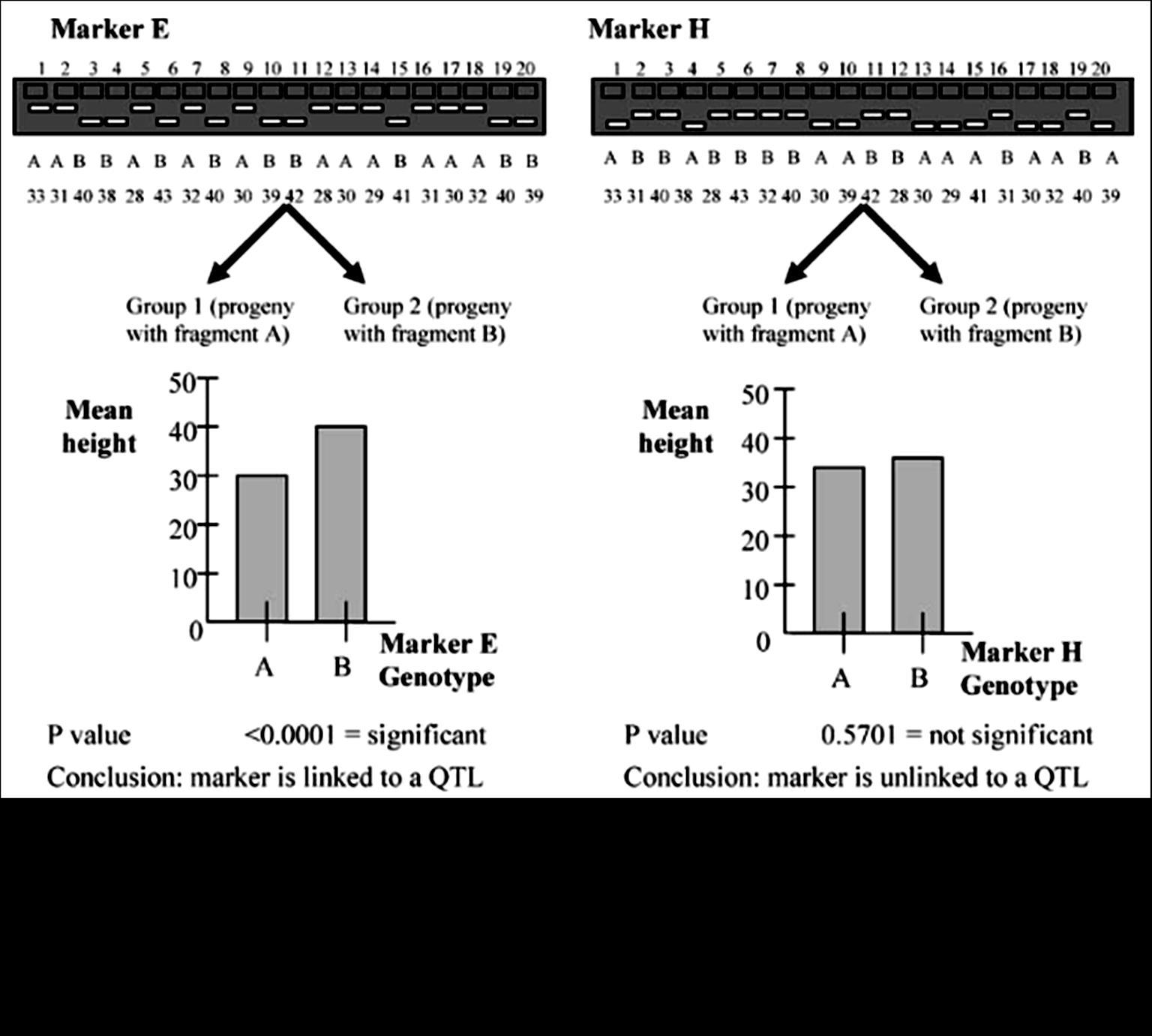

8.8.3.1 QTL mapping [Advanced]
Identifying a gene or QTL within a plant genome is like finding the proverbial needle in a haystack. How- ever, QTL analysis can be used to divide the haystack in manageable piles and sys-
tematically search them. In simple terms, QTL analysis is based on the principle of detecting an association between phenotype and the genotype of markers. Markers are used to partition the mapping population into different genotypic groups based on the presence or absence of a particular marker locus and to determine whether significant differences exist between groups with respect to the trait being measured, figure 8.75 A. A significant difference between phenotypic means of the groups (either 2 or 3), depending on the marker system and type of population, indicates that the marker locus being used topartition the mapping population is linked to a QTL controlling the trait
A logical question that may be asked at this point is ‘why does a significant P value obtained for differences between mean trait values indicate linkage between marker and QTL?’ The answer is due to recombination. The closer a marker is from a QTL, the lower the chance of recombination occurring between marker and QTL.
Crazygenes
Deletion in CCR5geneimpartsresistance to HIV, Plague,smallpox in WestEuropeanpopulation
Therefore, the QTL and marker will be usually be inherited together in the progeny, and the mean of the group with the tightly-linked marker will be significantly different (P < 0.05) to the mean of the group without the marker. When a marker is loosely-linked or unlinked to a QTL, there is independent segregation of the marker and QTL. In this situation, there will be no significant difference between means of the genotype groups based on the presence or absence of the loosely- linked marker. Unlinked markers located far apart or on different chromosomes to the QTL are randomly inherited with the QTL; therefore, no significant differences between means of the genotype groups will be detected.
Methods to detect QTLs: Three widely-used methods for detecting QTLs are singlemarker analysis, simple interval mapping and composite interval mapping, Single-marker analysis (also ‘singleanalysis’) is the simplest method for detecting QTLs associated with single markers. The statistical methods used for single-marker analysis include t-tests, analysis of variance (ANOVA) and linear regression.
Linear regression is most commonly used because the coefficient of determination (R2 ) from the marker explains the phenotypic variation arising from the QTL linked to the marker. This method does not
Figure9.Diagramindicatingtightandlooselinkagebetweenmarkerand crosses)occursbetween population.Markersthataretightlythe
aQTL(Marker H) are

require a complete linkage map and can be performed with basic statistical software programs. However, the major disadvantage with this method is that the further a QTL is from a marker, the less likely it will be detected. This is because recombination may occur between the marker and the QTL. This causes the magnitude of the effect of a QTL to be underestimated. The use of a large number of segregating DNA markers covering the entire genome (usually at intervals less than 15 cM) is useful.
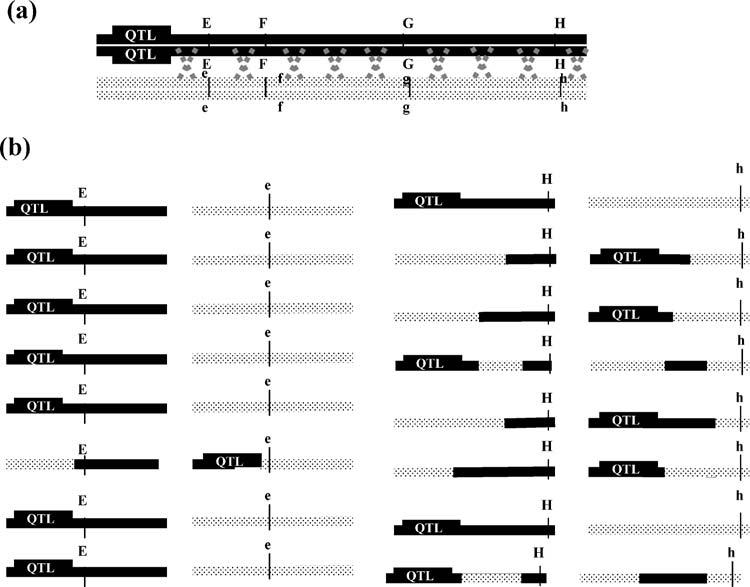
36.Therearetwoalleles,Aand a, governingbeaksize in apopulation: AA and Aa individual's beaksare1.5cm while aa individual'sare1cm. If thefrequency of theAallele in apopulation is 0.25,what is thepopulationmeanignoringenvironmentaleffects?
37. If additivevarianceforacharacter is 3andtotalphenotypicvariance is 6,then what is the heritability of thecharacter?
38.Imagine we areselectingporcupines to makethempricklier.Theheritability of prickliness is 0.75.Averagepricklinessbeforeselection is 100(standarddeviation is also100).Theaverage prickliness of theporcupineswhoproducethenextgeneration is 108.Whatwilltheprickliness of thenextgenerationbe?
39 Is thestatementtrue:Polygenictraitsaretraitsthatareonlyaffected by geneticinfluences?
40.Whatcolorwouldyouexpectforthehulls of a wheat plantheterozygousforallthreehull colorloci?
1. Any changes in the DNA of an organism which are unique and heritable, are termed as Mutations
2. Mutations can arise from chemical modifications, ionizing radiations or by spontaneous errors


3. Mutations are the sole reason for evolution and will remain the most important part of life

4. Mutations does not occur to cause disease, rather it helps us to survive by creating newer alleles
5. Mutation is a chance event and it may have positive or negative effect on organism and on community.

6. There are various types of mutations: Point mutation, Frame shift mutations, large lesions, suppressor mutations…etc.
7. Mutation can be reverted back to normal state, and hence they are reversible

8. Mutations are hypothesized to occur spontaneously and there has been no strong proof of conscious mutation by an organism.
9. Mutations from somatic cells are never passed onto next generation
10. Mutation rate is fixed for a population and it depends on the level of mutagen and advantages of mutated allele
11. Mutation can be suppressed by another mutation indicating that mutation is not a permanent change
12. Except mutation , nothing is permanent in life
We evolve in nature by acquiring new characters and capabilities. We find large diversity of life forms on earth. Why and how it may be happening? …It is due to some kind of changes in genetic sequence which altered the protein coded and there by function of an organism. These changes were passed on from one generation to the next, thus making them Heritable. We call such spontaneous heritable change in genetic level as Mutations.
Mutations can happen at any time and in any cell. The changes can be visible or it may be too minor change. These mutations may be lethal or beneficial to the organism.
broad categories of mutations:
mutations
mutations
mutation occurs in somatic cells
Some mutations can be turned on or off, depending on the environmental condition tional mutations.
Agents which cause Mutation
Mutagens
Spontaneous mutations
sudden occurrence in any point of time
Induced mutations
Mutations are not bad, as such. They are just periodic / untimey changes in DNA structure resulting in new sequences and chromosome structures. Mutation is the sole source for genetic diversity and evolution. With no mutation, no life can ever tolerate the changing environment with no mutation. It does not always result in diseases or problems.
Molecular basis of mutation
All kinds of mutations are due to changes in genetic material. Alterations in DNA sequences are the molecular causes of mutations. The gene sequences can be caused by deletions, insertions or rearrangement of DNA sequences in the genome.
Any mutation resulting from change in single base pair or any mutation resulting from single locus change in chromosome map
There are few important types of Point mutations, briefed below. Base substitution
simplest form of mutation
Nucleotide pair in a duplex DNA is replaced with a different nucleotide pair.
Purineis replaced by a purine base Purineis replaced by a pyrimidine
replaced by a pyrimidine
replaced by a purine

Mutation changes one codon into another
But codes for sameamino acid
Codon for one amino acid is changed into a codon for another aminoacid
Changes the aminoacid coded
Codon for one amino acid is changed into a terminationcodon
Addition or deletion of base in theORF
Causes premature termination of Causes changes inbase pairs
Some mutagens have similar structure as nitrogen bases of DNA. We call them as Base analogs. They interfere the DNA replication process ( they confuse the DNA polymerase enzyme)
5-bromo uracil
it is a base analog of Thymidine.
It acts in enol form to pair with Guanine.
In its keto form, it pairs with Adenine.
It causes G.C conversions
2-amino purine (2-AP)
It is a base analog of adenine
It can pair potentially with thymine
When protonated, it can pair with cytosine.
It causes G.C conversions
Some mutagens are not incorporated into the DNA
They can , simply alter a base, causing specific mispairing
Ethylmethanesulfonate(EMS)
It adds ethyl group to many positions on all 4 bases.

It adds methyl group to many positions on

4 bases
In general addition of O-6 alkyl guanine
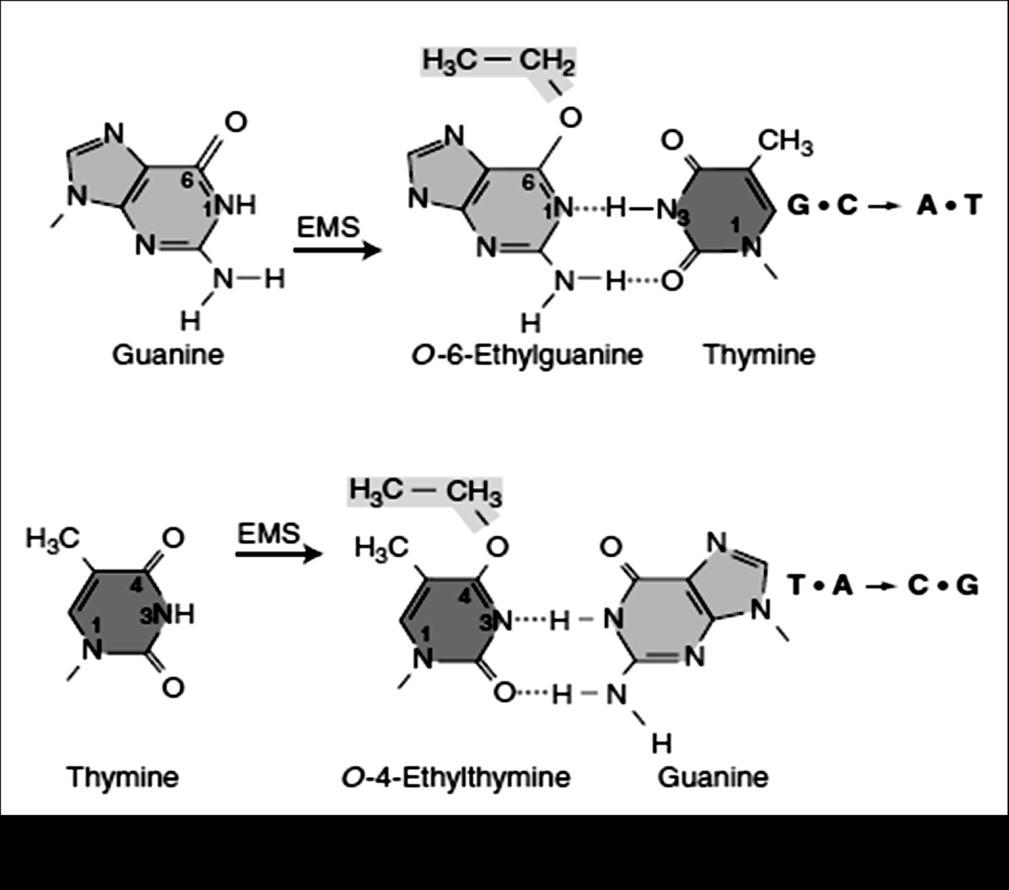
very potent
It is seen in most mutation cases.
Important DNA modifiers
Includes, proflavin, acridine orange, ICR compounds
They get themselves between 2 bases, called intercalation
Alter the DNA structure and therby modifying the gene expression pattern
They causeframe shift mutation, and there by changing thegene expression.
UV or any ionizing radiation causes, the base pairing patterns in DNA. These radiations are high energy photons and thus give high amount of energy, to the bases. These higher energy forms start adverse bonding with neighboring rupting the DNA helical structure. Usually UV radiation causes cancer of skin. rays generate a number of different alterations in DNA. We call them as products Most common errors are products.( thymine dimers)
A gene consists of promoter elements, enhancers, mRNA coding regions and terminator elements. The Reading frame consists of all the necessary codes for the proteins, any alterations in the sequence has direct effect on the protein coded. Any alteration in the Open Reading Frame [ORF] is called Frame shift Mutation. changes the “
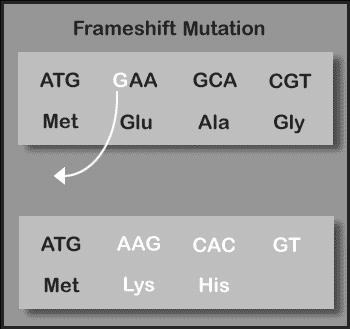

Original: he fat cat ate the wee rat.
Frame Shift (“a” added): The fat caa tat eth ewe era t.
Usually these are caused by Intercalating agents.
Spontaneous mutations arise suddenly in a cell, may be due to any errors in repli-
the readingframe,
the shift or change in
aminoacidcoded,this
cation or any chemical reactions happening in the cellular environment. These mutations are the major reason for evolution of new genes, proteins and new species. Let us try to understand some experiments, which help us to find out a mutation is a spontaneous mutation or not.
This was developed by two scientists Luria and Delbruck in 1943. They inoculated 20 small cultures, each with few cells Incubated until there were 108 cells/mm A larger culture vessel was maintained separately with 108 cells/mm 20 individual cultures and 20 samples from large culture were plated in the presence of Bacteriophage. Interestingly, the 20 small culture cells showed high degree of resistance to bacteriophage, than the cells from larger culture vessel. Thusthey show fluctuation in resistance to phage. Now, we have 2 possible explanations for this.
1.Mutation was induced by the phage. 2.Mutation occurred randomly before the phage infected.
If mutation was induced by the phage, the cultures form small and large stocks should have shown similar resistance, and there would be no fluctuation. Hence 1st is not the reason .Then it must be that mutation has occurred before the phage infection. Let us try tofind it out with the help of one more technique Replica plating Replica plating (Figure 8.83)
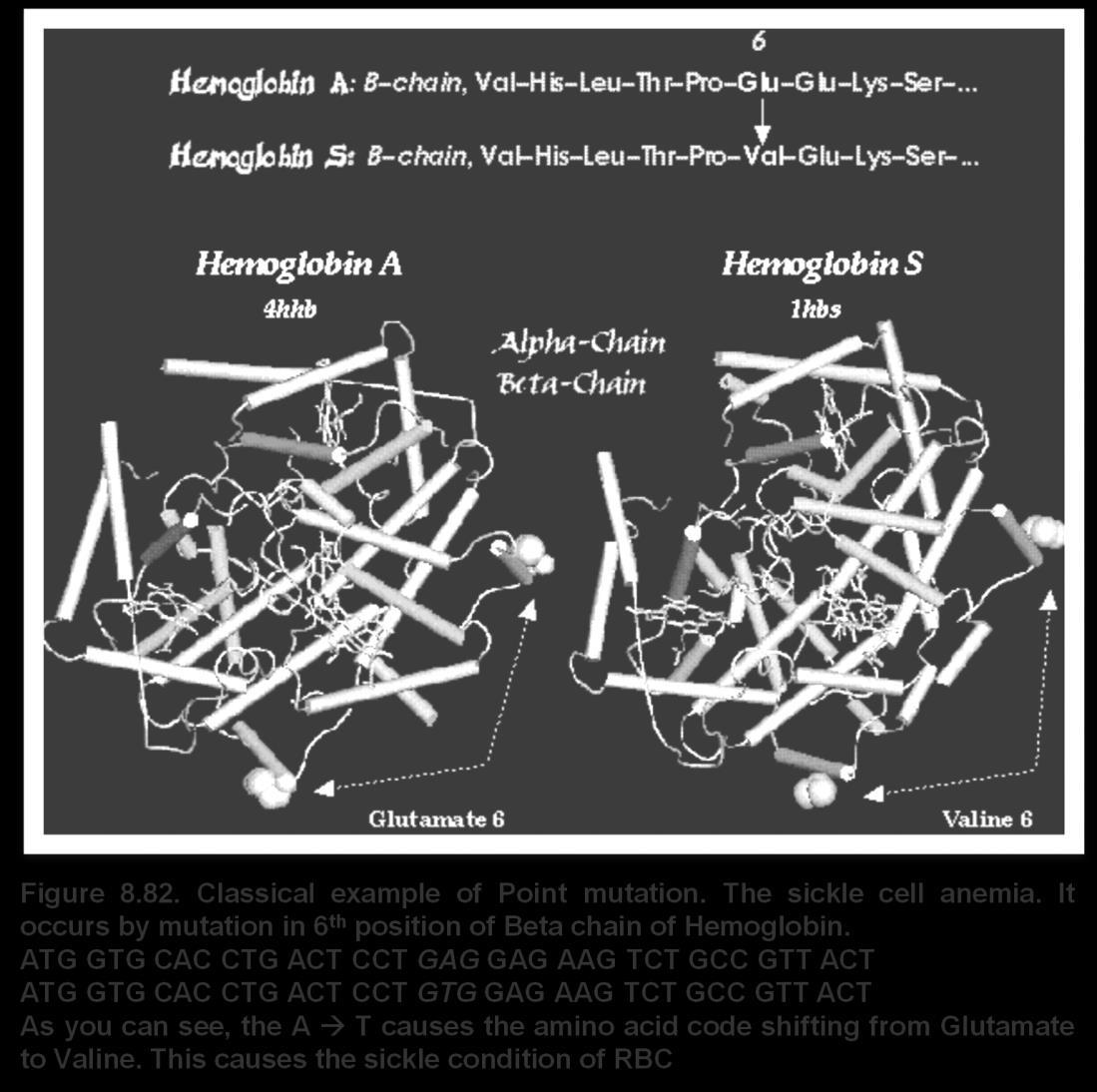
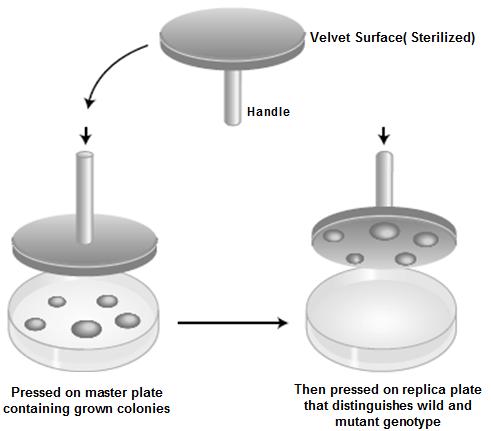
This technique was developed by Joshua and Esther in 1952
A bacterial culture is plated on non-selective medium ( medium with no bacteriophage)
We call that plate as
Master plate
A sterile velvet cloth was pressed down surface of the master plate.
Few cells get attached to the cloth.
velvet cloth
pressed onto
medium containing

After inoculation, the cells which are resistant to bacteriophage will grow on the medium. In this way, we can locate the mutants which are already present before the bacteriophage infected. Mechanism of Spontaneous mutations: Two most common type of mutations, 1. Depurination and 2 Deamination. Depurination has been dealt in above section. Deaminatioon involves removal of amino group from the nucleotide bases. Deamination of cytosine yield Uracil. Deamination of 5-methylcytosine generates Thymine It can convert G.C A.T pair.
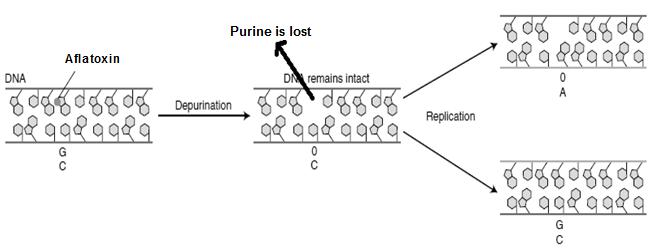

It is a powerful carcinogen from fungal origin position. It causes breakage of N-Glycisidic bond between the base and the sugar molecule. Creating Apurinic site (the base is removed) It induces G.C T.A transversions
Oxidative Damage
Most important and most potent mutation event
It is caused by
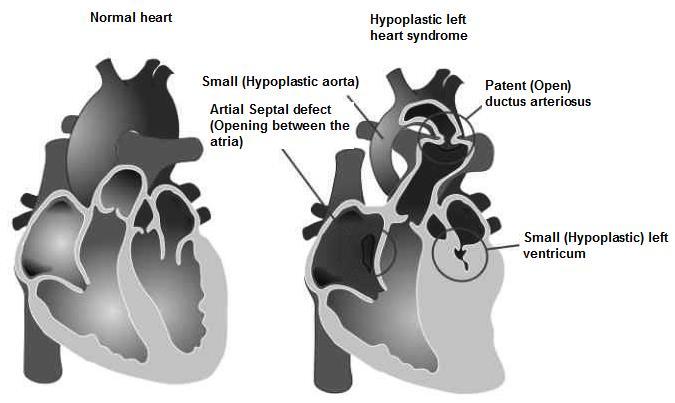
Superoxide radicals (O
Hydrogen peroxide (H
Hydroxyl radicals (OH They are produced as bi-products of normal metabolic pathway. They can oxidative
damage to DNA, resulting in change in structure of bases. Below figure 8.86 shows th products of Oxidative damage. Spontaneous mutations can be generated by several different processes. Spontaneous lesons and replicative errors generate most of base –substitutions, base pair changes. Spontaneous mutations in humans – Trinucleotide repeat diseases. Many diseases have been found today, which occur by simple repeats in human genome. Repeats are found in constant number in a
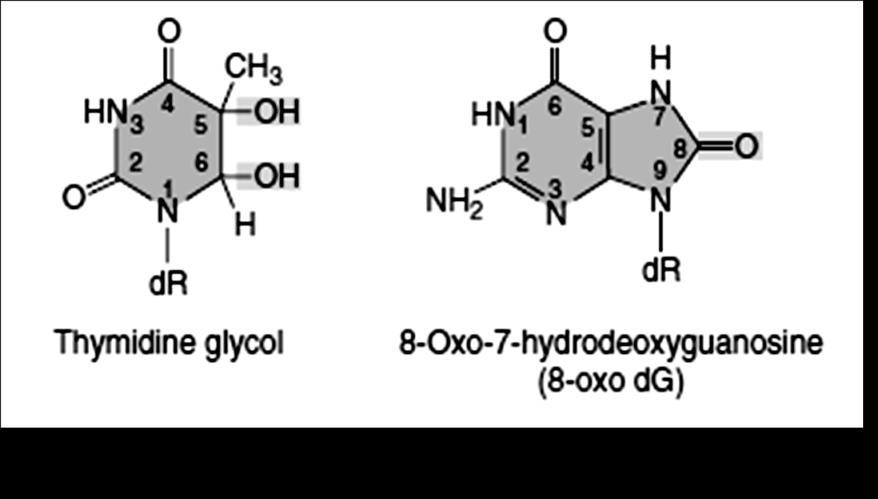

Find the notes of Insertional mutagenesis, in Unit XIII, under section 13.9


Chromosomal alterations

by
effect
by
which results in increase or decrease in number of one particular
results by improper duplication

can be called as Monosomy, Nullisomy, Disomy, Tetrasomy, Pentasomy, Trisomy..etc
disjunction
Improper separation of homologous chromosomes during gamete formations, results in aneuploidy
In human the Aneuploidy have serious impact resulting many genetic disorders
learned
mutations
genetic
basics of chromosome
chromosome
fibers
centro-
shapes
bases change.
behavior
this section
characters
telomeres
chromosomes
chromosome
end.
end of chromosome.
Chromosomal rearrangements
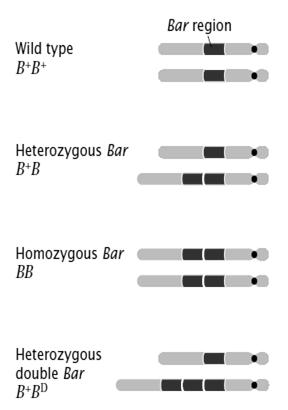
are the mutations that change the structure of chromosomes.
basic rearrangements
A mutation in which part of a chromosome has been doubled.
duplication occurs immediately next to original sequence tandem duplication

duplicated part is located at some distance from original mutation, displaced duplication
duplication is inverted, reverse duplication
DNA replication, the duplicated part forms a loop, as shown in the diagram.
This type of chromosomal
Loss of a segment of a chromosome


not undergo reverse mutation.
cause recessive genes on undeleted chromosome to ge expressed
causes imbalances in gene products
During meiosis, the non-deleted portion loops out and is clearly visible.
the deletion includes centromere, the chromosome will not segregate in meiosis and will usually be lost.
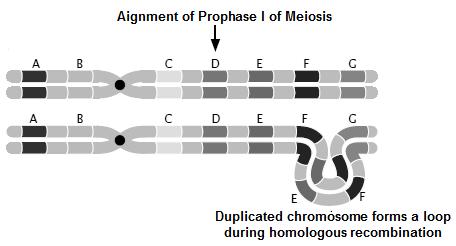
deletions are lethal in homozygous state.
barphenotype on drosophila eye..is aresult of X-linked duplication. As thenumber of duplicated genesincrease in chromosome,the phenotypedecreases. Thus duplication causesproblems in phenotypic expression.
Heterozygous for deletion

normal and other deleted homologous chromosome) create imbalance in amount of gene products.
a chromosome has R allele, dominant over r allele.
of the homologous chromosome has ‘R’ allele, and the other has ‘r’ allele.
deletion occurs in the chromosome containing R allele,
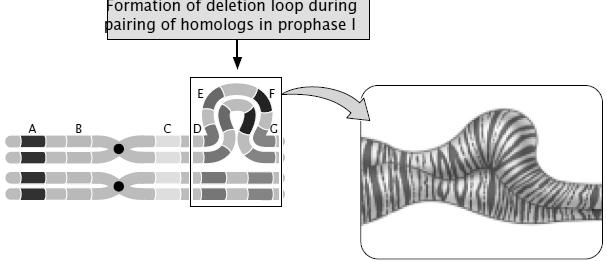
the cell is only left with ‘r’ allele.
that case,
allele
no other dominant control
Thus by deletion,
1. Cri du chat syndrome
its phenotype
recessive allele, became dominant
f part of short arm of chromosome
Cat like cry
Widely spaced eyes
Small head
Round face
Mental retardation
2. Wolf-Hirschhorn syndrome
in short arm of chromosome
Seizures
mental and growth retardations
Chromosome segments get inverted 180 degrees and reattaches
the inversion includes the centromere

the inversion do not include centromere:
results in No gain or loss of genetic material.
the order gets altered.
may break a gene into
genes are controlled by their positions,
by inversion position gets altered
the gene expression pattern
changes.
effect
affects the crossing over event.
8.10.4 Translocation
involves the movement of genetic material between non-homologous chromosomes or within same chromosomes.
parts,
the gene function.
Translocation does not mean crossing over.
crossing over gene exchange takes place between homologous chromosomes;
But in
Non reciprocal
material moves from one chromosomes to another without
reciprocal exchange
non-homologous
translocations
2 way exchanges.
call them as Reciprocal translocations
is special case of Reciprocal
becomes joined to a common centromere
generates a metacentric chromosome with 2 long arms
small chromosome
formed with 2 short arms
short chromosome
leads to decrease
chromosome
Most eukaryotes are diploid(2n) for most of their cell cycles, possessing 2 sets of chromosomes.
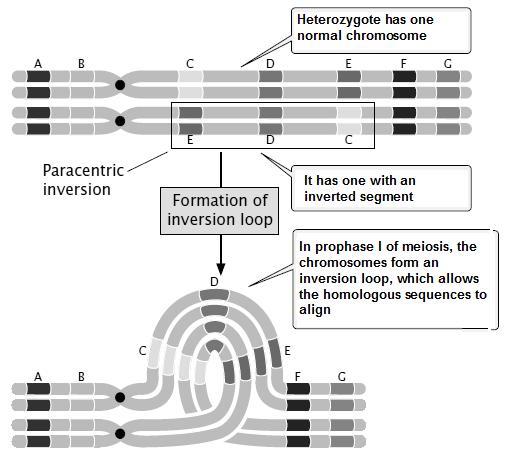
Sometimes, whole set of chromosome fail to separate in meiosis or mitosis.
leads to increase in entire set of chromosome
polyploidy condition
Triploids (3n) tetraploids(4n) pentaploids(5n)…etc
is the most common phenomenon in plants
is the major mechanism by which new plant species has
wheat, oats, cotton , potatoes, sugarcane…etc are polyploids
in
There are 2 types of polyploidy:
Autopolyploidy Auto polyploids
the sets of chromosomes are derived from same species of ancestors.
of all chromosomes
mitosis(2n)
tetrploids(4n)
crossing auto

diplod plant producing
haploid gamete.
This type of chromosomal rearrangement involves changes in chromosome number.
There are 2 kinds of Aneuploidy
Changes in number of individual chromosome
in number of chromosome sets due to non-disjunction during meiosis I , especially maternal
Aneuploidies disturb the delicate balance of gene products in cells by changing the chromosome number.
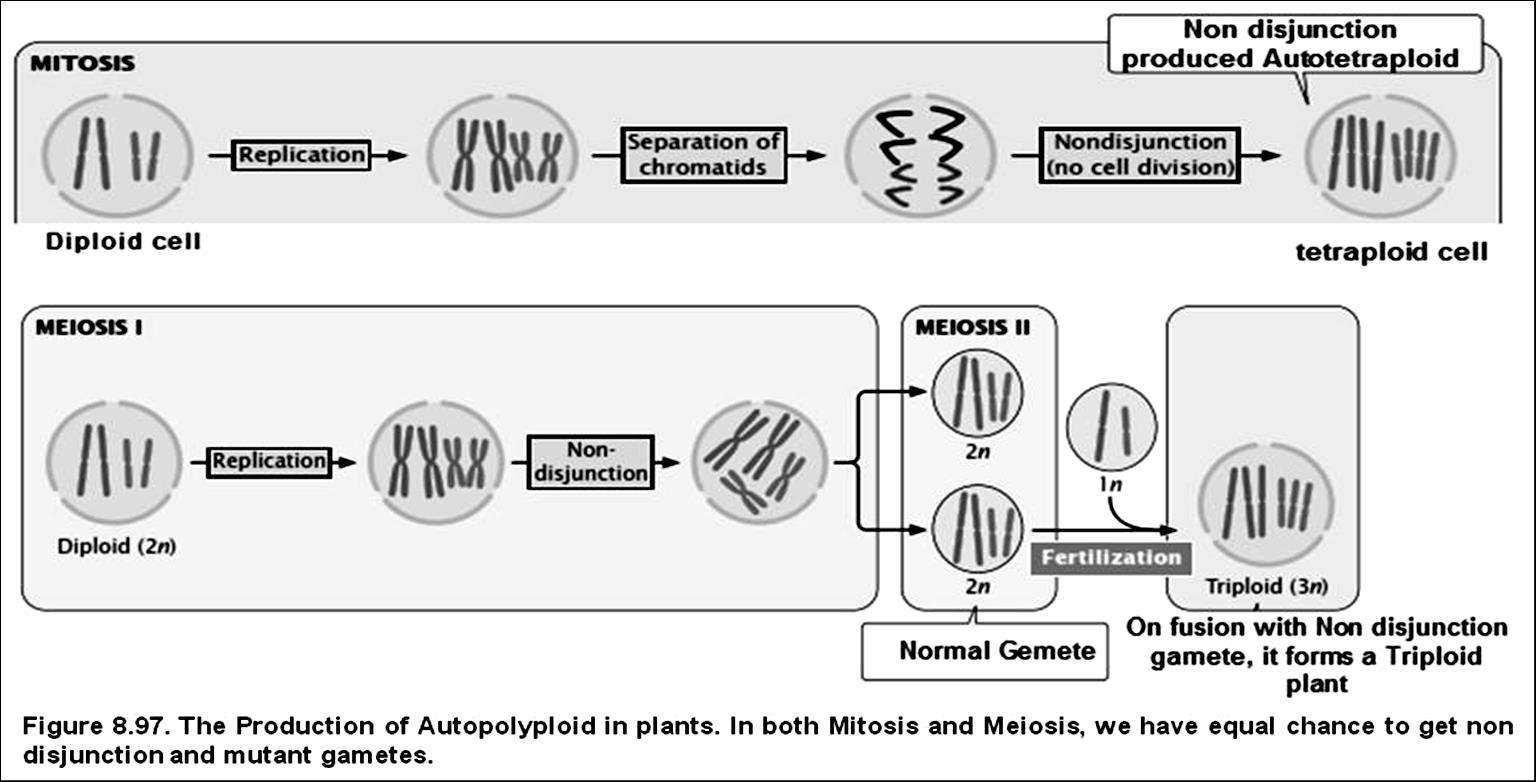
In humans, the most common aneuploidies are trisomies, which represent about 0.3% of all live births. Trisomies are characterized by the presence of one additional chromosome, bringing the total chromosome number to 47. With few exceptions, trisomies do not appear to be compatible with life. In fact, trisomies represent about 35% of spontaneous abortions
Many sex chromosome mosaics have been detected for example X/XX, X/XY, XX/XY and XXX/XXXXY
Since most embryos with chromosome abnormalities will fail to implant or will result in mis-

carriage, the frequency of aneuploidy in IVF embryos is much higher than the number of babies born with chromosome abnormalities. For example, a 40-yearold woman has approximately a 1 in 66 (1.5%) risk of having a liveborn child with a chromosome problem. At the time of IVF for a 40-year-old woman, approximately 60% of embryos will be abnormal, the majority of which would result in failed implantation or miscarriage.
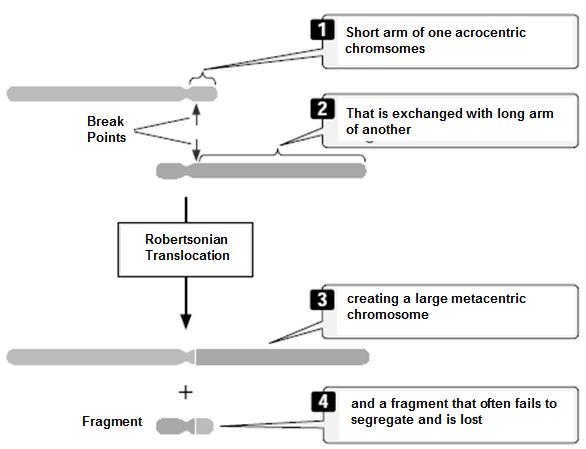
Figure8.98.NonDisjunction.
1.Acellwith2n=4cells,undergoingmeiosis.
2.Chromosomalseparation,reductiondivisionduringAnaphase uniform

3.MeiosisIcomplete,2cellswithhalfnumberchromosomesexpected, onecellandthe othergetsonly1.
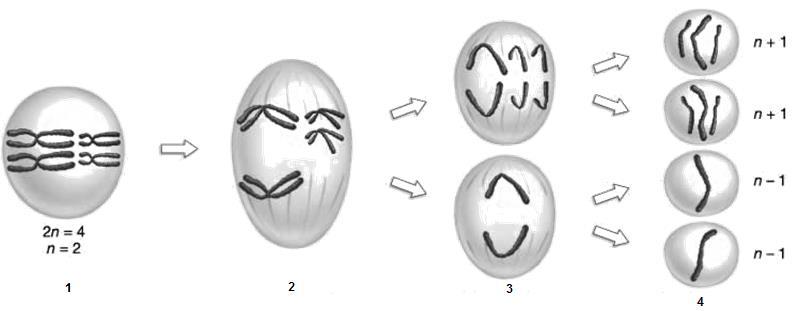
4.Meiosis II complete,4gametesformed,2cellswith
Figure8.99.TheHumanchromosome2containsa Robertisoniantranslocationthat is not present in chimps,gorillas or orangutan.G-bandingrevealsthat aRobertisiantranslocation in ahumanancestor switchedthelongandshortarms of the2acrocentric chromosomesthatarestillfound in theother3 primates.Thistranslocationcreatedthelarge metacentrichumanchromosome2.
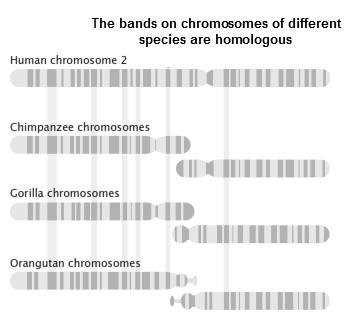
.Mosaicismforthesex chromosomesproducesaGynandromorphs. ThisXX/X0Gynandromorphfruitflycarriesone wildtypeXchromosomeandoneX chromosomewithrecessiveallelesforwhite eyes andminiaturewings.Theledtside of the flyhasanormalfemalephenotype,becausethe cellsare XX andtherecessivealleles onone X chromosomearemasked by thepresence of wildtypealleles on theother. The rightside of theflyhasamalephenotypewithwhiteeysand miniaturewing,becausethecellsaremissing thewildtypeXchromosome(areXO)allowing thewhiteandminiaturealleles to be expressed.
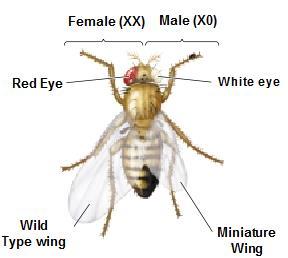
Nondisjunction in a mitotic division may generate patches of cells in which every cell has a chromosome abnormality and other patches in which every cell has a normal Karyotypes. This type of Nondisjunction leads to regions of tissue with different chromosome constitutions, a condition known as Mosaicism
Growing evidence suggests that Mosaicism is relatively common.
Only about 50% of those diagnosed with Turner syndrome have the 45,X Karyotypes (presence of a single X chromosome) in all their cells, most others are mosaics, possessing some 45,X cells and some normal 46,XX cells.
A few may even be mosaics for two or more types of abnormal Karyotypes.
The 45,X/46,XX mosaic usually arises when an X chromosome is lost soon after fertilization in an XX embryo.
Fruit fly that is XX/XO mosaics (O designates the absence of a homologous chromosome; XO means the cell has a single X chromosome and no Y chromosome) develop a mixture of male and female traits, because the presence of two X chromosomes in fruit flies produces female traits and the presence of a single X chromosome produces male traits independently in each cell during development.
Those cells that are XX express female traits; those that are XY express male traits. Such sexual mosaics are called
Normally, X-linked recessive genes are masked in heterozygous females but, in XX/XO mosaics, any X-linked recessive genes present in the cells with a single X chromosome be expressed.


Results from hybridization between 2 species
The polyploidy plant will have chromosome sets arising from 2 or more species.
Cabbage + radish
This plant has roots of cabbage and leaves of radish
Usually polyploids are sterile and but may possess some beneficial characters.

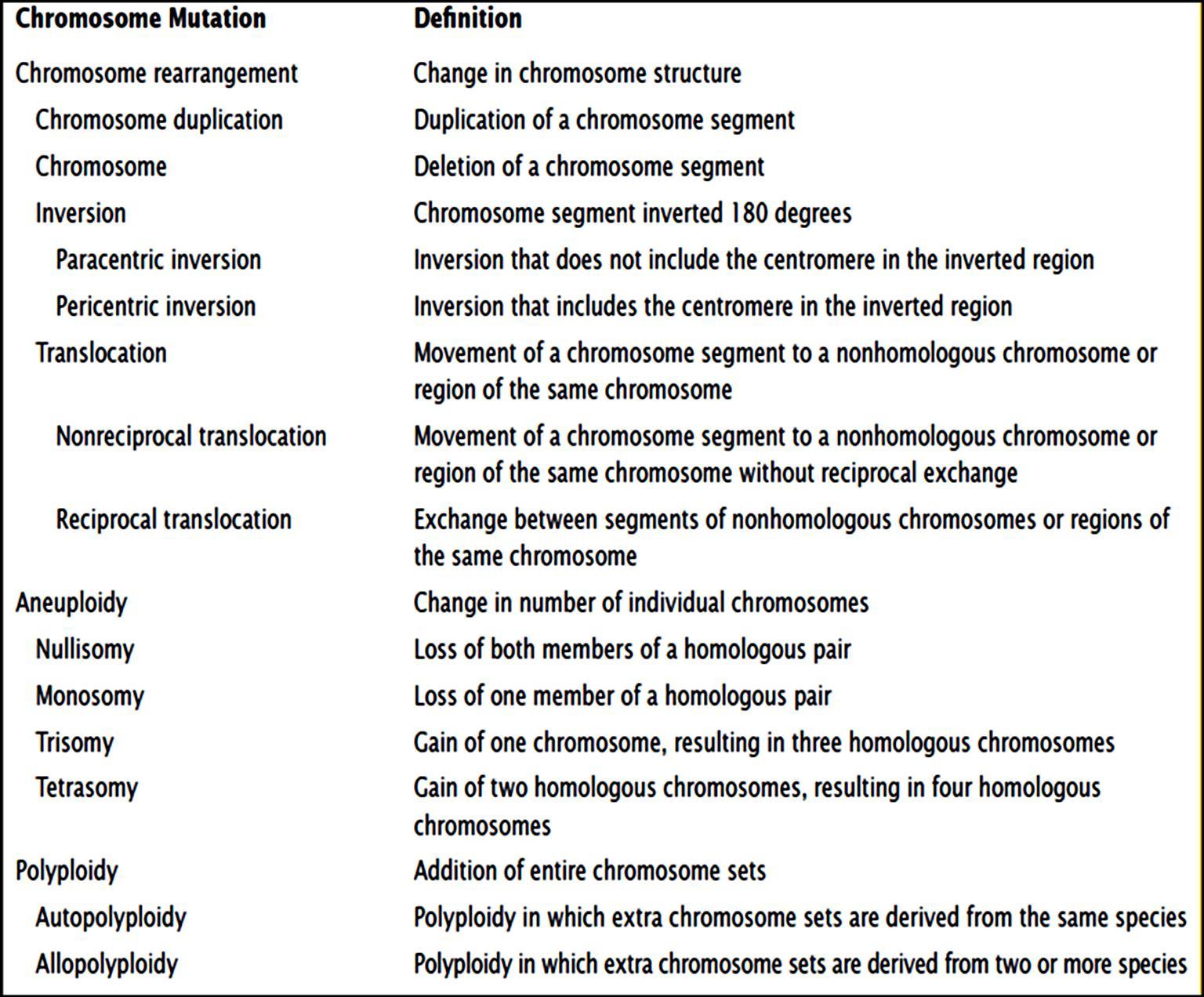
Find the note under Unit 3.1, Recombination.

1. What do we inherit from parents? Genotype or Phenotype?
Genotype. Phenotype is developed with respect to environment
2. How many different combination s of maternal and paternal chromosomes can be packaged in gamete mad by an organism with a diploid number 8?
64 [8x8]
3. What was the reason for Mendel not to observe deviation in Pea plant experiment?
Because he choose characters from different chromosomes, so no question of linkage
4. What do you think, do we find Mendelian inheritance in Human beings too??
Yes, we do have many characters. Viz, Ability to taste phenylthiocarbamide (dominant), Ability to smell (bitter almond-like) hydrogen cyanide (recessive), Albinism (recessive)
Brachydactyly (shortness of fingers and toes)...etc., Wet (dominant) or dry (recessive) earwax, dry is found mostly in Asians and Native Americans.
5. How a dominant gene actually suppresses the recessive gene?
It does not suppress as such, but expresses itself in such high level that the expression of recessive gene is not visible in phenotype
6. Complementation experiment reveals that gene is a divisible, yet distinguished entity. Explain Complementation test revels that the genes can be divided and again reuniting them restores the character. But we cannot have all possible combination, genes are unique but divisible.
7. In what ways the complementation test useful?

It will help us
8. What is actual structure of gene?
Gene actual structure is still a mystery, only thing we know is the
9. In terms of Dominance and recessive, what actually meant in molecular terms?
The gene with strong promoter and enhancer elements becomes the dominant and the one with weak promoters becomes recessive. This makes the dominant gene express often in high expressivity.
10.The gene which has been silenced from many centuries, can we call it as Pseudo gene?
No, it can just be masked, but not mutated.
11. If gene A and B are linked by 10cM distance, what might be the probability of obtaining the homozygous gamete for gene A and B?
Chance of recombination is 10%, so 90& of time homozygous combination AB and ab will appear.
12. How many linkage groups are there in human male?
24 [all autosomes +X+Y]
13. Why there is difference between the genotype and the people showing its phenotype?
Having a genotype does not matter, but the expression matters. The phenotype is mediated by the environment and not just by genes.
14.What do you think, the parents donate genes to offspring in equal number?
Yes, donation of genes is equal but expression pattern in unequal
15.Whenever, we say dominant genes, recessive genes, what do we refer to at molecular level?
We refer to its operon structure and promoter sequences similarity to consensus sequence
16. How would you predict whether a character is actually genetically coded or environmentally coded?
By pedigree analysis
17. Linked genes always stay linked, is the statement true? If yes/No what is your explanations?
No, they do not linkage is always partial.
18.Linkage and recombination are the property of genes and chromosomes, does it depend on presence or absence of introns?
No, intron has nothing to do with linkage, but yes it may pose problems as it increases the distance between the genes.
19.How many linkage groups, human female has?
23
20. If there were complete linkage between Mendel tall and dwarf genes, how the F2 ratio would appear?
No recombinants and ration would have been 1:1 :: Tall: Dwarf
21. What is the difference between the maternal effect and Maternal inheritance?
Maternal effect : The cytoplasmic mRNA and proteins given by mother egg cell, affects initial growth of embryo.
Maternal inheritance: The mitochondrial genes affect the phenotype of the offspring.
22. What are the factors on which the inheritance of characters depend on?
It depends on the environment, selection, fitness of the gene.
23. If maternal inheritance and maternal effect has intense influence on the organism, then why don’t we all resemble our mother?
They only shape initial growth and most of the genes are concentrated in nucleus, only few ATP synthesis genes are in mitochondria
24.What do you think when might be the mRNA for egg formation in females is produced in female body?
When she is in her mother’s womb, 12
25.What would be the reason for severe side effects in our body under overdose of antibacterial medicine?
It affects the mitochondria as it shares features with prokaryotes.
26. Why microbial gene recombination is called as Horizontal mode of gene transfer?
There is no new cell formation, but the gene gets transferred in the same generation bacteria, not to next generation.
27. How would you differentiate whether 2 bacteria produced a wild type by recombination or by mutation?
Separate the bacteria colonies and check for phenotypes, if the wild type is still restored it must be mutation, if they fail to produce wild type it must have been by recombination.
28.F+ donates plasmid to F- cells, in the light of evolution, can you explain the helping nature of F+, even though there is no gain to F+?
F+ tries to increase its colony to survive, so it needs more bacteria of its kind. It’s not helping, it’s again the selfish nature for survival.
29.If at all the genes to be transferred into F- cells, how long the conjugation should continue? 100 min
30.Does the conjugation only takes up the good genes? No, it takes lace randomly, non-selective
31. Is the dominant inheritance and dominant allele same? No, dominant inheritance, the gene flow in all generation with environmental advantages. Dominant gene is ju
32. If pedigree analysis, had to involve the linked genes, what statistical tool comes into picture? Jeninfer tool
33. Pedigree analysis in standard conditions ignores some of the genetic phenomenon in large. Can you name a few of them?
Penetrance, expressivity, genomic imprinting and all epistatic interactions, Pleotropy, Phenocopy..etc

34.How do you think the pedigree of Co-dominant alleles would behave?
Heterozygotes will show the expected phenotype and both homozygotes normal
35.Can we find out with the help of pedigree location of genes on chromosomes? No, we can’t we only can conclude on autosomes or on allosomes
36. There are two alleles, A and a, governing beak size in a population: AA and Aa individual's beaks are 1.5cm while aa individuals are 1cm. If the frequency of the A allele in a population is 0.25, what is the population mean ignoring environmental effects?
1.22 cm
37. If additive variance for a character is 3 and total phenotypic variance is 6, then what is the heritability of the character?
0.5
38.Imagine we are selecting porcupines to make them pricklier. The heritability of prickliness is 0.75. Average prickliness before selection is 100 (standard deviation is also 100). The average prickliness of the porcupines who produce the next generation is 108. What will the prickliness of the next generation be? 106
39.Is the statement true: Polygenic traits are traits that are only affected by genetic influences?
No, they can also be effected by environment and no. of alleles present
40. What color would you expect for the hulls of a wheat plant heterozygous for all three hull color loci? Medium red
41.Nothing is permanent in world except mutation. Justify?
Yes, mutation is always happening and nothing can stop it, if mutation stops, no life can sustain for long 42.Mutations are always occurring in response to environment, is the statement true? No, mutations can happen spontaneously without any requirement of agent.
43.If mutations are so easily done, then why not we change overtime rapidly?
Every system has its own repair-damage control system to check the mutation rate.
44.Name any 5 important disorders arising due to mutations.
22q11.2 deletion syndrome; Angelman syndrome; Canavan disease; Charcot Tooth disease; Color blindness
45.How would you predict the mutation in operator region affects the gene expressions?
Mutation in operator gene would make either the gene over active or total loss in activity.
46.What would be the effect of chromosomal mutation on sperm shape?
Lethal, the sperm is very sensitive to chromosomal changes and any change in sperm structure would lead it infertile.
47.Name any laboratory chemical which can cause severe chromosomal aberrations? Colchicine
48.What would be the effect of polyploidy in humans?
Human cell [except few] with polyploidy cell will not be viable.
49.Chromosomal mutation and gene mutations, which is more lethal?

Chromosomal mutation, because gene mutation can be repaired by our system, but chromosomal mutation, we can’t repair.
50. As chromosome number increases mutation rate increases, is the statement true?
Yes it is, more no. of genes and more room for mutation.

1. How many different chromosome configurations can occur following meiosis I if three different pairs of chromosomes are present (n = 3)?
a. 4 b. 8 c. 16 d. 9
2. If two parents, both heterozygous carriers of the Autosomal recessive gene causing cystic fibrosis, have five children, what is the probability that exactly three will be normal?
a. 0.5 b. 0.25 c. 0.1 d. 0.8
3. If 2n = 18, what will be the no. of chromosomes in a triploid organism? a.18 b.27 c. 19 d.17
4. If a single bacteriophage infects one E. coli cell present on a lawn of bacteria and, upon lyses, yields 200 viable viruses, how many phages will exist in a single plaque if three lytic cycles occur?
a. 200 b. 400 c. 2004 d. 2003
5. Inversions are said to “suppress crossing over.” Is this terminology technically correct?

a. Yes , true b. No, its false
c. Cant determi
d. May be , Iam not sure
6. Consider an inherited trait in Drosophila with the following characteristics: (1) It is transmitted from father to son to grandson. (2) It is not expressed in females. (3) It is not transmitted through females. Is the trait likely to result from extra nuclear inheritance?
a.Yes
b. No c. It is Autosomal paternal d. Its Autosomal maternal
7. The rate of mitochondrial DNA evolution is given as 1 nucleotide change per mitochondrial lineage every 1500 to 3000 years. Given that human mitochondrial DNA is approximately 16,500 nucleotides, what is the rate of change per nucleotide per million years?
a. 2-4% nucleotide changes / year
b. 2% nucleotide changes / year
c. 4% nucleotide changes / year
d. 1-10% nucleotide changes / year

8. Two strains of maize exhibit male sterility. One is cytoplasmically inherited; the other is Mendelian. What will be the result of crosses involving un related strains and Mendelian male sterile plant?
a. No male sterile plant or 50% male sterile plant
b. All male sterile plant
c. No male sterile plant
d. 50% male sterile and 50% female sterile as Mendelism involves allele segregation.
9. A temperate bacteriophage has the gene order “a b c d e f g h”, whereas the order of genes in the prophage present in the bacterial chromosome is “g h a b c d e f”. What information does this give you about the location of the attachment site in the phage?
a. Between a and b
b. Between b and c
c. Between g and f
d. None of the above
10. Consider the following Hfr ×F- cross. Hfr genotype: a+ b+ c+ strF-genotype: a- b- c- str-r
The order of gene transfer is a b c, with a transferred at 9 minutes, b at 11 minutes, c at 30 minutes, and str-s at 40 minutes. Recombinants are selected by plating on a medium that lacks particular nutrients and contains streptomycin. Can I conclude that the a+ str-r colonies > c+ str-r colonies.
a. True b. False c. May be
d. Can’t determine
11. Down syndrome occurs in 1 in 700 individuals. If in a year, 42 million people are born, how many in that are expected t have Down syndrome?
a. 6000
b. 60000
c. 600000 d. 15000
12. The chromosomal arrangement in nullisomy is a.2n-1
b. n-1
c. 2n+1 d. n+1
13. What determines the maximum number of different alleles that can exist for a given gene?
a. Gene promoter region
b. Gene location on chromosome
c. Gene size
d. Type of organism
14. Genetic disease Alkaptonurea results in accumulation of which compound in urine?
a. Urea
b. Uric acid
c. Homogentisic acid
d. Ureco gentisic acid
15. One of the possible cure for Phenylketonuria is
a. Stop intake of phenyl alanine
b. Stop intake of tyrosine

c. Control intake of phenylalanine

d. Control the intake of tyrosine
16. What is the gene order on the bacterial chromosome:
Hfr strain Markers donates
1 DCBA
2 AGFE
3 BCDE
4 FGAB
a. ABCDEFG
b. ABDCFEG
c. BACDGFE d. EFGABDC
17. If 2 loci are 10 map units apart, what proportion of the meiotic events will contain a single crossover in the region between these 2 loci, assuming no multiple cross over occurs?
a.20%
b.10%
c. 5% d.2%
18. Genes a and b are 20 mU apart, in a test cross involving a+b+/a+b+ as one of the parent, how many gametes will be of a+a/b+b genotype?
a. 100% b.20%
c. 10% d.0%
19. In Davis U tube experiment, addition of DNAase enzyme, did not stop colony formation and growth. This indicates
a. Transduction is not occurring b. Transformation is not occurring c. Conjugation is not occurring d. Enzyme is not working.
20. In genetic anticipation
a. We can sense the disease before it occurs b. Person can anticipate the disease occurrence c. Time of disease onset decreases and severity increases as it is passed in generations d. None of the above
21. In incomplete dominance
a. Dominant gene is suppressed b. Recessive gene is dominating c. Both the gene products are expressed d. Both the gene products give a new kind of phenotype.
22. Pedigree chart will enable us to understand
a. The nature of the gene
b. The nature of the disease
c. The disease flow pattern d. The disease allele segregation in child
23. What will be the approximate effective population size in a panmitic population of 440 with 400 females and 40 polygamous males?
a. 363
b. 133 c. 663 d. 267
24. A mother of blood group 0 has a group A child. The father could be of blood type
a. A or B or O. b. A only.
c. A or B. d. A or AB only.
25. Aneuploid females with only one
a. Criduchat
b. Klinefelter syndrome c. Down syndrome. d. Turner syndrome.
26. A mechanism that can cause a gene to move from one linkage group to another is
a. Crossing over. b. Inversion. c. Translocation. d. Duplication.
27. The following is the biochemical pathway for purple pigment production in flowers of sweet pea:
Recessive mutation of either gene A or B leads to the formation of white flowers. A cross is made between two parents with the genotype: AaBb x aabb. Considering that the two genes are not linked, the phenotypes of the expected progenies are a. Purple: 7 white. b. 3 white: 1 purple.
c. 1 purple: 1 white. d. purple: 6 light purple: 1 white

28. The main reason for albinism is
a. Exposure to sun b. Mutation in DNA repair enzymes

c. Mutation in Tyrosine pathway d. Mutation in Tryptophan pathway
29. One of the two chromosomes that determines an individual’s gender is known as a(n)
a. autosome b. centromere
c. sex chromosome d. X-linked trait
30. A chart that shows the relationships within a family is a(n)
a. karyotype b. pedigree
c. nondisjunction d. promoter
31. What is an error in meiosis in which homologous chromosomes fail to separate?
a. point mutation
b. karyotype
c. nondisjunction

d. pedigree
32. The most common form of severly affected people who are having more than 1000 repeats of the CTG triplets are called a. Fragile X syndrome
b. Myotonic dystrophy
c. Retinoblastoma
d. Acute Myelogeous leukemia
33. The genetic change that occurs in more than 1% of the population
a. Polymorphisms
b. Monotheism
c. Frameshift mutations d. Inverted repeats
34. A point mutation
a. changes a single base pair. b. deletes at least 10 base pairs. c.inverts a section o d. is always fatal.
35. Banti and Bubly are planning a family, but since each has a brother with sickle cell anemia, they are concerned about their child. Neither banti , bubly nor their parents have the disease, in that case what would you predict about their children?
a. There is very little chance that any of their children will have sickle cell disease b. That all of their children will have sickle cell disease c. That one out of four of their children could be expected for the disease d. That it is possible that none of their children will have disease but blood test on them will be required to make sure
36. Which of the following is true about aneuploidy?
a. Inversion b. 2n+1 c. All aneuploid individuals die before birth d.4n
37. In a population at large, there is a rare disease that affects one person in a thousand. A diagnostic test for the disease is 99% accurate. This means that there is a 1% chance that the test wrongly identifies a healthy person as having the disease. Suppose you take the test and the result is positive. Then the chance that you indeed have the disease is about a.9% b.1% c. 50% d. 0.1%
38. In a family, both husband and wife have blood group ‘A’. The first child born to them was found to have ‘O’ blood group. What is the probability that their next child will have blood group ‘A’? a. 0.0 b. 0.25 c. 0.50 d. 0.75
39. What is the probability to obtain the plant with AabbCcDd genotype from parent AABBCCDD and aaBbCcDd is a. ½ b. 1/16
c.
40. What do you call an organism that is capable of making its own food store?
a. Heterotroph
b. Photo autotroph
c. Autotroph
d. Chemolithotroph
41. A frameshift mutation
a. Deletes an entire chromosome.
b. Replaces one codon with a different one. c. Can move from place to place in the genome. d. Removes a base pair, offs
42. Robertsonian translocation occurs between
a. Acrocentric human chromosomes
b. Telocentric chromosomes
c. Metacentric chromosomes
d. Only between sex chromosomes
43. Which of the following is true With regard to the DNA structures and genetic codon?
I. Adenine (A) and guanine (G) are purine bases
II. guanine (G) always pairs with thymidine (T) and adenine (A) with cytosine (C)
III. each DNA strands hav
IV. there are 64 possible codons
V. Each amino acids may be coded by more than one codon

a. true; II- False; III- True ; IV b. true; II- Tr c. False; II- False; III- True ; IV d. true; II- False; III- True ; IV
44. In tomato, the following genes are located on chromosome 3: +Tall plants + normal leaves + smooth fruit ‘d’ dwarf plants ‘m’ mottled leaves ‘p’ pubescent fruit Result of the cross: +++ / dmp x dmp/dmp were a. +mp / d++ b. ++p / dm+ c. +m+ / d+p d. +++/dmp
45. In Mendel’s experiment the Round seeds character (RR) is completely dominant over the Wrinkled seed character (ss). If the character for the height were in completely dominant such that TT are tall, Tt are intermediate and tt are short, what would be the phenotypes resulting from crossing a Round seeded short (SStt) plant to a Wrinkled seeded, tall (ssTT) plant?
a. All the progeny would be Round seeded and tall b. ½ would be Round seeded and intermediate height/ ½ would be Round seeded and tall c. All the progeny would be Round seeded and short d. All the progeny would be s Round seeded and intermediate height
46. Those mutations that arise in the absence of known mutagen are known
a. Induced mutations
b. Fused mutations
c. Spontaneous mutation d. None of the above
47. Which of the following descriptions of chromosomes is not correctly matched
a. Metacentric chromosome arms are almost equal in size.
b. Submetacentric — chromosome arms are slightly different in size.
c. Acrocentric — chromosome arms are identical in size.
d. Telocentric there is only one chromosome arm.
48. A snail of the species Limnaea peregra in which the shell coils to the left undergoes self-fertilization, and the entire progeny coil to the right. What is the genotype of the mother?
a. s+s
b. ss
c. both
d. Can’t determine
a.
49. Consider a heritable autosomal disease with an incidence in the population of 1 per thousand. On average, individuals with the disease have 80% as many children as the population average. What mutation rate would be required to maintain the observed incidence of the disease in the population if the disease is dominant?
a. 0.01
b. 0.001
c. 0.0001 d. 0.00001
50. In some sheep, the presence of horns is produced by an Autosomal allele that is dominant in males and recessive in females. A horned female is crossed with a hornless male. One of the resulting F1 females is crossed with a hornless male. What proportion of the male and female
a. ½ of male and ½ of females
b. ¼ of males and ½ of females
c. ¼ of males and none of females
d. ½ of males and none of females
51. Research has shown that a particular eye defect is represented in a family pedigree as follows:
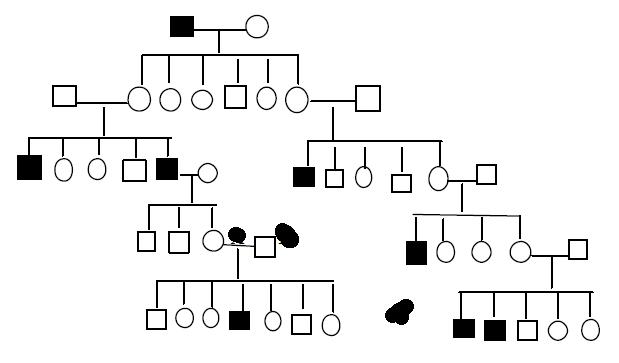
On the basis of this data, which of the following mechanisms of inheritance are most POSSIBLE?
a. Autosomal dominant, b. Autosomal recessive, c. sex-linked dominant, d. Y-linked
52. A rare dominant mutation expressed at birth was studied in humans. Records showed that six cases were discovered in 40,000 live births. Family histories revealed that in two cases, the mutation was already present in one of the parents. Calculate the spontaneous mutation rate for this mutation.

a. 2x10-4
b. 3x10-5
c. 5x 10-5
d. 5 x 10+5
53. Presented here are hypothetical findings from studies of heterokaryons formed from seven human xeroderma pigmentosum cell strains:

These data are measurements of the occurrence or nonoccurrence of unscheduled DNA synthesis in the fused heterokaryon. None of the strains alone shows any unscheduled DNA synthesis. How many different complementation groups are revealed based on these data?

a. 1 b. 2 c. 3 d. 4
54. Microsatellites are currently exploited as markers for paternity testing. A sample paternity test is shown in the following table in which ten microsatell and an alleged father. The name of the microsatellite locus is given in the lefthand column, and the genotype of each individual is recorded as the number of repeats he or she carries at that locus. For example, at locus D9S302, the mother carries 30 repeats on one of her chromosomes and 31 on the other. In cases where an individual carries the same number of repeats on both chromosomes, only a single number is recorded. (Some of the numbers are followed by a decimal point, for example, 20.2, to indicate a partial repeat in addition to the complete repeats.) Assuming that these markers are inherited in a simple Mendelian fashion; can the alleged father be excluded as the source of the sperm that produced the child?
a. No, he is the real father as evident form chart; he has given every allele to the child.
b.yes, he is not the real father as he has not given any of the alleles c. It not confirmed, need additional data about the frequency of markers in population, hence cant draw any definite line d. Both a and c
55. Consider the two very limited unrelated pedigrees shown here. Which is the modes of inheritance can be absolutely ruled out in each case?
a. X-linked recessive b. X-linked dominant c. Autosomal recessive d. Autosomal dominant
56. Tay–Sachs disease (TSD) is an inborn error of metabolism that results in death, often by the age of 2. You are a genetic counselor interviewing a phenotypically normal couple who tell you the male had a female first cousin (on his father’s side) who died from TSD and the female had a maternal uncle with TSD. There are no other known cases in either of the families, and none of the matings have been between related individuals. Assume that this trait is very rare. Calculate the probability that both the male and female are carriers for TSD.
a. ¼
b. 1/8
c. ½

d. 3/4
57. Dentinogenesis imperfecta is a tooth disorder involving the production of dentin sialophosphoprotein, a bone-like component of the protective middle layer of teeth. The trait is inherited as an autosomal dominant allele located on chromosome 4 in humans and occurs in about 1 in 6000 to 8000 people. Assume that a man with dentinogenesis imperfecta, whose father had the disease but whose mother had normal teeth, married a woman with normal teeth. They have six children. What is the probability that their first child will be a male with dentinogenesis imperfecta?
a. 5/16
b. 2/9
c. 5/64
d. 2/64
58. In a certain plant, fruit is either red or yellow, and fruit shape is either oval or long. Red and oval are the dominant traits. Two plants, both heterozygous for these traits, were test crossed, with the following results. Determine the location of the genes relative to one another.

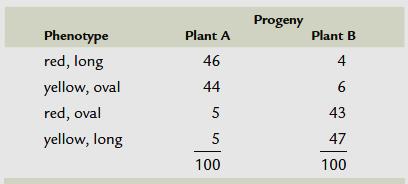
a. 20 mu b. mu c. 40 mu d. 15 mu
59. In laboratory class, a genetics student was assigned to study an unknown mutation in Drosophila that had a whitish eye. He crossed females from his true-breeding mutant stock to wild type (brick-red-eyed) males, recovering all wild-type F1 flies. In the F2 generation, the following offspring were recovered in the following proportions: The student was stumped until the instructor suggested that perhaps the whitish eye in the original stock was the result of homozygosity for a mutation causing brown eyes and a mutation causing bright red eyes, After much thought, the student was able to analyze the data, explain the results, and learn several things about the location of the two genes relative to one another. One key to his understanding was that crossing over occurs in Drosophila females but not in males. Based on his analysis, what did the student learn about the two genes?
a. Genes are complete linkage in males
b. Genes are partially linked in males c. Genes are completely linked in females d. Genes are not present in males
60. Time mapping is performed in a cross involving the genes his, leu, mal, and xyl. The recipient cells were auxotrophic for all four genes. After 25 minutes, mating was interrupted with the following results in recipient cells. Diagram the positions of these genes relative to the origin (O) of the F factor and to one another.

1. 90% were xyl+
2. 80% were mal+
3. 20% were his+
4. none were leu+
a.xyl-mal-his-leu
b.Xyl-mal-leu-his
c. Leu-mal-xyl-his
d. Mal-o-his-leu-
61. In Bacillus subtilis, linkage analysis of two mutant genes affecting the synthesis of two amino acids, tryptophan (trp2-) and tyrosine (tyr1-), was performed using transformation. What can you conclude from the following dat
a. The genes are quite closely linked together.
b. The genes are not linked together.
c. The genes are completely linked together.
d. No inference can be drawn.
62. The Xg cell-surface antigen is coded for by a gene located on the X chromosome. No equivalent gene exists on the Y chromosome. Two codominant alleles of this gene have been identified: Xg1 and Xg2. A woman of genotype Xg2/Xg2 bears children with a man of genotype Xg1/Y, and they produce a son with Klinefelter syndrome of genotype Xg1/Xg2/Y. Using proper genetic terminology, briefly explain how this individual was generated. In which parent and in which meiotic division did the mistake occur
a. Non disjunction in Meiosis-I of father
b. Non disjunction in meosis-II of father
c. Non-disjunction in meiosis-I of mot d. Non disjunction in meiosis-II of mother
63. In Drosophila, the X chromosomes may become attached to one another (XX) such that they always segregate together. Some flies thus contain a set of attached X chromosomes plus a Y chromosome. What sex would such a fly be?
a. Male b. Femal
c. Both the above
d. No sex
64. Individuals with Down syndrome, while suffering congenital defects with tendencies toward
a. Respiratory disorders
b. Bone disorders

c. Brain disorders
d. Hand disorders
65. What is true about the following pedigree?
a. Disease is paternally inherited
b. Genes are on X chromosomes

c. Disease is patenally inherited and the gene is on Y chromosome
d. Disease is maternally inherited
66. In a cross of Limnaea, the snail contributing the eggs was dextral but of unknown genotype. Both the genotype and the phenotype of the other snail are unknown. All F1 offspring exhibited dextral coiling. Ten of the F1 snails were allowed to undergo self-fertilization. One-half produced only dextrally coiled offspring, whereas the other half produced only sinistrally coiled offspring. What were the genotypes of the original parents?
a. DD and DD
b. DD and dd c. DD and Dd d. dd and dd
67. Tay Sachs disease is caused by loss- tations in a gene on chromosome 15 that encodes a lysosomal enzyme. Tay Sachs is inherited as an autosomal recessive condition. Among Ashkenazi Jews of Central European ancestry, about 1 in 3600 children is born with the disease. What fraction of the individuals in this population are carriers?
a. 1 in 30
b. 1 in 300 c. 2 in 30 d. 1 in 3000
68. In Drosophila, Lyra (Ly) and Stubble (Sb) are dominant mutations located at loci 40 and 58, respectively, on chromosome III. A recessive mutation with bright red eyes was discovered and shown also to be on chromosome III. A map is obtained by crossing a female who is heterozygous for all three mutations to a male homozygous for the bright red mutation (which we refer to here as br). The data in the table are generated. Determine the location of the br mutation on chromosome III.
a. Gene br is in center
b. Gene ‘br’ is at right corner
c. Gene ‘br’ is at left corner

d. Both b and c are possible
69. Two chemically induced mutants, xand y,are treated with the following mutagens to see if revertants can be produced: 2-amino purine (2AP), 5-bromouracil (5BU), acridine dye (AC), hydroxylamine (HA), and ethylmethanesulfonate (EMS). In the following table, + means revertants and - means no revertants. For each mutation, determine the probable base change that occurred to change the wildtype to the mutant.
a. x+ x is ATGC ; y+ to y is GC AT

b. x+ x is GCAT ; y+ to y is GC AT
c. x+ x is ATGC ; y+ to y is AT GC
d. x+ x is GCTA ; y+ to y is GC AT
70. You have isolated a set of five yeast mutants that form dark red colonies instead of the usual white colonies of wild-type yeast. You cross each of the mutants to a wild-type haploid strain and obtain the results shown below. Which of the following mutants are dominant to wild type?
a. Mutant 1
b. Mutant 2
c. Mutant 3
d. Mutant 4
71. Consider a hypothetical insect species that has red eyes. Imagine mutations in two different unlinked genes that can, in certain combinations, block the formation of
red eye pigment yielding mutants with white eyes. In principle, there are two different possible arrangements for two biochemical steps responsible for the formatio of red eye p gene would block the formation of red pigment. Alternatively, the two genes could act in parallel such that mutations in both genes would be required to block the formation of red pigment. When mutant phenotype is not reverted back to wild type we can conclude that the
a. Mutant genes belong to same complementation group
b. Mutant genes belong to different complementation group
c. Mutant genes do complement each other d. Mutant genes did not revert back to wild type by gene conversion]
72. Genes that control coat color in mammals represent some of the best early examples of genes with multiple alleles exhibiting different phenotypes. A classic example is the A gene in hamsters. Three of the known alleles of this gene are A, a- and a+. Each supplies a different amount of black pigment to the coat hair. The phenotypes of three different homozygous strains are as follows
AA a-a- Sepia fur a+a+ Albino fur
By series of crosses, we assumed that Black is dominant over both a- and a+ .When we crosses the albino (a+a+) and sepia (a-a-), we got all heterozygous hamsters which were NOT black. What this result suggests about the 2 alleles?
a. The a- and a+ are the different alleles of different genes
b. The a- and a+ are the different alleles of same genes
c. The a- and a+ are the same alleles of different genes
d. The a- and a+ are the same alleles of same genes
73. The following pedigree shows the segregation of 2 different recessive traits.
Assuming that both traits are due to linked autosomal genes that are 10 cM apart, calculate the probability that the indicated child will have both recessive traits.
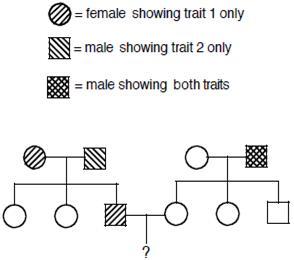
a. 0.5
b. 0.4
c. 0.225
d. 0.65
75. Barney is an alien. On his ship, hidden in the Stata building, you find alien bacteria that metabolize wood. You call this species A. termiticus, and call your original strain BLT (for “Barney’s little termiticus”). You subject a sample of BLT to mutagens, and isolate a new strain that no longer metabolizes wood. You conclude that you have succeeded in disrupting at least one gene necessary to metabolize wood. You call the mutant strain M. You mix a sample of M with a sample of heat-killed wild type BLT, and the resulting strain metabolizes wood. You summarize your data in the following table
Strains
BLT
Heat-killed BLT
M
Heat-killed BLT +M Yes
a. What do you conclude about the genetics of the bacteria?
b. The heat killed + Mbacteria underwent mutation
c. The heat killed +M bacteria acquired genes by transformation
d. The heat killed +M bacteria acquired genes by conjugation
e. The BLT strain lost its property due to loss of genetic material which would in fact code for enzyme to digest the wood.
74. You have isolated a mutant strain A. termiticus which can metabolize wood. You would like to know its genetic material. You start by breaking some A. termiticus cells open to determine their molecular composition. You find that they contain various small molecules,
carbohydrates, lipids, and two other macromolecules, A and B. In order to determine which macromolecule is the carrier of genetic information, you include test tubes where you treat the sample of the heat-killed BLT with either an agent that destroys macromolecule A (A-ase) or macromolecule B (B-ase). You find the following result

BLT
Heat killed BLT
M
Heat-killed BLT +M
A ase treated heat
+M
B ase treated heat +M
Which molecule is the carrier of genetic information in A. termiticus?
a. A
b. B
c. Both d. Can’t be determined
75. Consider a portion of an autosome in Drosophila, which carries the following three recessive markers: black body (b–), purple eyes (pr–), and vestigial wings (vg–). The corresponding wild type alleles of each gene are designated b+, pr+, and vg+. A genetic map of this portion of the chromosome is shown here: “b” 6cM “pr” 13cM “vg” . What would be the distance between the “b” and “vg” if you measure the distance by 2 point cross?
a. 13 cM
b. 19 cM
c. More than 19 cM
d. Less than 19 cM
76. You have isolated two different yeast mutants that will not grow on medium that lacks arginine. You call these mutants arg1– and arg2–. Mating of either arg1– or arg2– to wild type produces diploids that can grow without arginine. Mating of arg1– to arg2– produces a diploid that also can grow on medium without arginine. What do these results tell you about the arg1– and arg2– mutations?
a. The two mutants are recessive to wild type and they complement each other
b. The two mutants are dominant to wild type and they complement each other
c. The two mutants are recessive to wild type and they do not complement each other
d. The two mutants are dominant to wild type and they do not complement each other
77. You have isolated two temperature-sensitive mutations in phage l. These phage mutants are called ts-1 and ts-2. Each mutant will form plaques at 30˚C but not at 42˚C. You cross ts 2 phage by coinfecting E. coli at the permissive temperature of 30˚C. When the resulting phage lysate is plated at 30˚C you count 105 plaques per ml of phage lysate, but when the same phage lysate is plated at 42˚C, there are 300 plaques per ml. What is the distance between the ts
a. 6 mU
b. 0.6 mU
c. 10 mU d. 50 mU
78. Wild type E. coli metabolizes the sugar lactose by expressing the enzyme ß-galactosidase. You have isolated a mutant that you call lac1, which cannot synthesize ß-galactosidase and cannot grow on lactose (Lac ). You have a wild type (Lac+) strain carrying a Tn5 insertion
known to be near several Lac genes on the E. coli chromosome. You grow P1 phage on this strain and use the resulting phage lysate to infect the lac1 strain, selecting for kanamycin resistance (Kanr). Among 100 Kanr transductants, you find that 82 are Lac and 18 are Lac+. Express the distance between Tn5 and
a.10%
b.18%
c. 22%
d. 28%
79. You are running a human assisted reproduction clinic and providing state- -the-art genetic diagnostic services. A married couple who already had a child with cystic fibrosis approach you because they wish to have another child, but only if they can be assured that the child will not have cystic fibrosis. You genotype the woman and discover that she is a heterozygote for Del508, the most common mutation causing cystic fibrosis. You suggest that the couple consider first polar body testing, in which several unfertilized oocytes (each with its first polar body) are retrieved from the woman, the first polar bodies are removed, and PCR tests are conducted on DNA from each of the first polar bodies. The couple agrees, and you obtain the following results:
PCR test of first polar body DNA for presence of Oocyte
#1
+
Why did the polar bodies for oocytes #1 and #5 test positive for both Del508 and the wild type sequence, while the polar bodies for the other oocytes do not?
a. Recombination between the Del508 and the wild type occurred in oocyte 1 and 5

b. Recombination between the Del508 and the wild type occurred in all other oocytes except 1 and 5
c. The genes Del508 and wild type were linked in Oocyte 1 and 5
d. The data is not sufficient to understand the molecular details, we need to map the genes to respective chromosomes, as sex chromosomes cause problems in determination of genes.
80. Assuming that both parent plants in the diagram below are homozygous, why would all of the f1 generation have yellow phenotypes?
a. Because yellow parent is homozygous
b. Because yellow parent is heterozygous
c. Because yellow parent is hemizygous
d. Because yellow is dominant to green
81. In manx cats, the gene (M) for tail-lessness is dominant such that M is a lethal allele, so MM kittens are not viable If you mate a large number of tail-less heterozygous (Mm) cats what kind of proheny we can expect?
a. All one tailed kitten for every two tail-less kittens.
b. All two tailed kitten for every one tail-less kittens.
c. ¾ the one tailed kitten and ¼ th tailed kittens.
d. Tailed and tail-less kittens in equal number
82. Most Border Collie dogs are black and white. The allele for this is B and the trait is autosomal dominant. Homozygous recessive dogs are chocolate and white. When a test cross was done with black and white dog, we got all black and white puppies, what does this result tell you about the genotype of the black and white parent?
a.BB
b.Bb
c. bb
d. None of the above
83. The following are human blood types, note that alleles “A” and “B” are co
recessive. What are the possible blood types of offspring of type A father and type B mother?
a. Only A
b. Only B
c. Only A and B
d. Any of the AB
84. We inherit from equal genes from both parents, is the statement true?
a.Yes
b. No c. Somewhat d. Don’t know
85. Neurofibromatosis type 1 is one of the most common Autosomal dominant disorders. A woman with neurofibromatosis type 1 has an unaffected partner. Which of the following is correct regarding their children?
a. The probability that each of their children will be affected is 1 in 4.
b. The probability that their son is affected is 1 in 4.
c. The probability that their third child will be affected if their first two children are affected is 1 in 2.
d. If their first child is affected then their second child will not be affected.
86. Retinoblastoma usually occurs as a sporadic event but in around 10% of cases there is a positive family history due to Autosomal dominant inheritance. In these families the penetrance is approximately 80% (= 0.8). A woman with retinoblastoma, whose father and
grandfather were also affected, has an unaffected partner with no relevant family history. Which of the following is correct regarding their children?

a. The probability that their first child will be affected is 0.25.
b. The probability that their first child will be affected is 0.4.
c. The probability that each of their children will be affected is 0.5.
d. The probability that each of their children will be affected is 0.4 X 0.5.
87. In this pedigree I 1 and II 2 have Becker muscular dystrophy which shows X-linked recessive inheritance. III 2 has oculocutaneous albinism which shows autosomal recessive inheritance with an incidence in the general population of 1 in 10 000. The disease status for both Becker muscular dystrophy and oculocutaneous albinism is unknown for IV 1 and IV 2. What is the probability that III 3 is a carrier of Becker muscular dystrophy?
a. 1/4
b. 2/3
c. 1/2
d. 1
88. Which of the following is true of a species that has a chromosome number of 2n=16?
a. The species is diploid with 32 chromosomes
b. There are 16 homologous pairs
c. During the S phase of the cell cycle there will be 32 separate chromosomes
d. A gamete from this species has 8 chromosomes.
89. Huntington disease is a dominant autosomal disease. Unfortunately, onset of symptoms usually occurs after a person has reproduced. What are the chances that a heterozygous person with the disease will produce a child with the disease if mated with a normal homo-
zygous person?
a.0%
b. 100%
c. 25% d.50%
e.75%
90. Theoretically, if a female hemophiliac (recessive sex-linked gene) and a normal male had children, what percentage of their male offspring would be expected to have hemophilia?
a. 33.3%
b.25%
c. 50% d. 100% e.9%
91. Based on the following recombination frequencies between genes on the same chromosome, determine the correct order of the genes on the chromosome. X-A 25% E-A 50% M-A 30% X-E 25% M-E 80%

a. M X E A
b. M A E X
c. E M A X
d. E X A M
92. In yeast, 1 cM of genetic distance corresponds to 3,500 base pairs of physical distance. Using this data, can you guess the no.of bases in the DNA element of a chromosome?
a. 1.75 x 10^5 bases
b. 17.5 x 10^5 bases
c. 3.5 x 10^5 bases
d. 3.5 x 10^4 bases
93. If a is linked to b, and b to c, and c tod, does it follow that a recombination experiment would detect linkage between a and d? a.Yes
b. No
c. May be d. Can’t be determined
94. Mice have 19 autosomes in their genome, each about the same size. If two autosomal genes are chosen randomly, what is the chance that they will be on the same chromosome?
a. 1/20
b. 1/9
c. 1/19
d. 1/371
95. Genes a and b are 20 cM apart. An a+ b+/a+ b+ individual was mated with an a b/a b individual. If the F1 was crossed to a b/a b individuals, what offspring would be expected, and in what proportions?
a. 40% a+ b+/a b, 40% a b/a b, 10% a+ b/a b, 10% a b+/a b
b. 50% a+ b+/a b, 50% a b/a b
c. 40% a+ b+/a b, 10% a b/a b, 40% a+ b/a b, 10% a b+/a b
d. 40% a+ b+/a b, 10% a b/a b, 10% a+ b/a b, 40% a b+/a b
96. A homozygous variety of maize with red leaves and normal seeds was crossed with another homozygous variety with green leaves and tassel seeds. The hybrids were then backcrossed to the green, tassel-seeded variety, and the following offspring were obtained: red,
normal 124; red, tassel 126; green, normal 125; green, tassel 123. What do you conclude of these results?
a. The genes for plant color and seed type linked and assort independently
b. The genes for plant color and seed type un linked
c. The genes for plant color is dominant over seed type
d. The genes for plant color and seed type linked

97. A phenotypically wild-type female fruit fly that was hetero-zygous for genes controlling body color and wing length was crossed to a homozygous mutant male with black body (allele b) and vestigial wings (allele vg). The cross produced the following progeny: gray body, normal wings 126; gray body, vestigial wings 24; black body, normal wings 26; black body, vestigial wings 124. What is the frequency of recombination between body color and wing length?
a. 15.7%
b. 16.7%
c. 20% d.25%
98. In Drosophila, the X-linked recessive mutations prune (pn) and garnet (g) recombine with a frequency of 0.4. Both of these mutations cause the eyes to be brown instead of dark red. Females homozygous for the pn mutation were crossed to males hemizygous for the g mutation, and the F1 daughters, all with dark red eyes, were crossed with their brown-eyed brothers. Predict the frequency of sons from this last cross that will have dark red eyes.
a.40%
b.20%
c. 10% d.5%
99. Two strains of maize, M1 and M2, are homozygous for four recessive mutations, a, b, c, and d, on one of the large chromosomes in the genome. Strain W1 is homozygous for the dominant alleles of these mutations. Hybrids produced by crossing M1 and W1 yield many different classes of recombinants, whereas hybrids produced by crossing M2 and W1 do not yield any recombinants at all. What is the difference between M1 and M2?
a. M2 carries an inversion that suppresses recombination in the chromosome. b. M1 carries an inversion that suppresses recombination in the chromosome.
c. M2 carries an inversion that induces recombination in the chromosome.
d. M1 carries an inversion that induces recombination in the chromosome.
100. The following pedigree shows the inheritance of X-linked color blindness and hemophilia in a family. What are the frequency of recombination in the children if II-2?
a.40%
b.50%
c. 30% d.25%


3' - end the end of a polynucleotide with a free (or phosphorylated) 3' - hydroxyl group. 5' - end -- the end of a polynucleotide with a free (or phosphorylated or capped) 5' - hydroxyl group; transcription/translation begins at this end.

known form of short limbed dwarfism characterized byanormaltrunksizewith disproportionally short arms and legs,and a disproportionally large head; autosomal dominant condition.
Advanced maternal age (age 35 at delivery) at increased risk for nondisjunction trisomy in foetus. Alcoholism characterizedbytheinabilityto control theconsumption of alcohol. Allele --analternativeformofagene;anyoneof several mutational forms of a gene. Alpha-fetoprotein (AFP) thefoetusintotheamnioticfluidandfromthere into the mother's bloodstream through the placenta.
Alu repetitive sequence persed repeated DNA sequence in the human genomeaccountingfor5%ofhumanDNA.Thename isderivedfromthefactthatthesesequencesare cleaved by therestriction endonuclease Alu. Amino acid sequence -- the linear order of the amino acids in a protein or peptide.
Amniocentesis prenatal diagnosis method using cells in the amniotic fluid to determine the number and kind of chromosomes of the fetus and, when indicated, perform biochemical studies. Amniocyte cells obtained by amniocentesis.
Amplification any process by which specific DNAsequencesarereplicateddisproportionately greater than their representation in the parent molecules.
Aneuploidy stateofhavingvariantchromosome number (too many or too few). (i.e. Down syndrome, Turner syndrome).
Angelman syndrome a condition characterized bysevere mental deficiency, developmental delay and growth deficiency, puppet-like gait and frequentlaughterunconnectedtoemotionsofhappi-
a condition caused by the premature closure of the sutures of the skull bones, resulting in an altered head shape, with webbed fingers and toes. Autosomal dominant. theplacementofsperm into a female reproductive tract or the mixing of male and female gametes by other than natural
aphotographicpictureshowing the position ofradioactive substances in tissues. a nuclear chromosome other than a virus whose host is a bactethecondensedsingleX-chromosome nucleiofsomaticcellsoffemalemammals.basepair a pair ofhydrogen-bonded nitrogenous bases (one purine and one pyrimidine) that join the component strands of the DNA double
Base sequence -- a partnership of organic bases foundinDNAandRNA;adenineformsabasepair withthymine(oruracil)andguaninewithcytosine in a double-stranded nucleic acid molecule. Baysian analysis -- a mathematical method to further refine recurrence risk taking into account other known factors.
Unit
Becker muscular dystrophy X-linked condition characterized by progressive muscle weakness and wasting; manifests later in life with progression less severe than Duchenne muscular dystrophy.
Carrier an individual heterozygous for a single recessive gene.
cDNA complementary DNA produced from a RNA templ DNA polymerase.
Centromere

spindle traction fibers attach during mitosis and meiosis; the position of the centromere determines whether the chromosome is considered an acrocentric, metacentric or telomeric chromosome.
Charcot-Marie Tooth disease characterized by degeneration of the motor and sensorynervesthatcontrolmovementandfeeling in the arm below the elbow and the leg below the knee; transmitted in autosomal dominant, autosomal recessive and X-linked forms.
Chorionic villus sampling diagnostic procedure involving removal of villi from the human chorion to obtain chromosomes andcellproductsfordiagnosisofdisordersinthe human embryo.
thethreadlikestructuresconsistingofchromatin andcarrygeneticinformationarrangedinalinear sequence.
Chromosome banding chromosomes so that bands appear in a unique pattern particular to the chromosome. Cleft lip/palate congenital conditionwithcleft lip alone, or with cleft palate; cause is thought to be multifactorial.
Clone genetically engineered replicas of DNA sequences.
Cloned DNA any DNA fragment that passively replicates in the host organism after it has been joined to a cloning vector.
Codon asequenceofthreenucleotidesinmRNA that specifies an amino acid. Consanguinity geneticrelationship.Consanguineous individuals have at least one common ancestor in the preceding few generations. anaminoacidchangethat does not affect significantly the function of the
genes physically close on a chromosomethatwhenactingtogetherexpressa conditioninvolving growth deficiency, significant developmental delay,anomaliesoftheextremitiesandacharacter-
plasmid vectors designed for cloning largefragmentsofeukaryoticDNA;thevectoris a plasmid into which phage lambda cohesive end
areas of multiple CG repeats in a chromosomal condition (monosomy 5p). Name comes from the distinctive mewing cry of affected infants; characterized by significant mental deficiency, low birthweight, failure to thrive and short stature; deletion of a small section of the short arm of
the exchange of genetic material between two paired chromosome during meiosis. Cystic fibrosis an autosomal recessive genetic condition of the exocrine glands, which causes thebodytoproduceexcessivelythick,stickymucusthatclogsthelungsandpancreas,interfering with breathing and digestion.
Cytogenetics -- thestudy of chromosomes.
Unit
Degenerate codon a codon that specifies the same amino acid as another codon.
Deletion the loss of a segment of the genetic material from a chromosome.
Deletion mapping the use of overlapping deletions to localize the position of an unknown gene on a chromosome or linkage map. Disease any deviation from the normal structure or function of any part, organ, or system of the body that is manifested by a characteristic set of symptoms and signs whose pathology and prognosis may be known or unknown.
DMD Duch
DNA fingerprint technique
todeterminedifferencesinaminoacidsequences between related proteins; relies upon the presence of a simple tandem-repetitive sequences that are scattered throughout the human genome.
DNA hybridization
binding specific segments of single-stranded (ss) DNA or RNA by base pairing to complementary sequences on ssDNA molecules that are trapped on a nitrocellulose filter.
DNA probe
isolate a gene, a gene product, or a protein.
DNA sequencing synthesis"
synthesized in vitro in such a way that it is radioactively labeled and the reaction terminates specifically at the position corresponding to a givenbase;the"chemical"method,ssDNAissubjected to several chemical cleavage protocols thatselectivelymakebreaksononesideofaparticular base.
DOE Department of Energy. Dominant--allelesthatdeterminethephenotype displayed in a heterozygote with another (recessive) allele.
Down syndrome atypeofmentaldeficiencydue totrisomy(threecopies)ofautosome21,atranslocation of 21 or mosaicism.
Duchenne/Becker muscular dystrophy the most common and severe form of muscular dystrophy;transmittedasanX-linkedtrait.X-linked recessive. Symptoms include onset at 2-5 years with difficulty with gait and stairs, enlarged calf muscles, progression to wheelchair by adoles-
conditions of short stature with adult height under 4'10" as adult, usually with normal intelligence and lifespan. Ehlers Danlos Syndrome connective tissue condition including problems with tendons, ligaments, skin, bones, cartilage, and membranes surrounding blood vessels and nerves. Symptoms include joint laxity, elastic skin, dislocations. Many forms: autosomal dominant, autosomal recessive, X-linked forms. neurologic condition involving repeated twisting and movement. Involves avariety of muscle groups. Intelligence not effected. Threeforms:childhood-autosomaldominant,au-
ethical, legal and social implications (of

anenzymethatbreakstheinterdiester bonds in a DNA molecule. the hemoglobin-containing cell
the study of fundamental principles fines values and determines moral duty and obligation.
Euchromatin the chromatin that shows the staining behavior characteristic of the majority of the chromosomal complement.
Eugenics -- the improvement of humanity by altering its genetic composition by encouraging breeding of those presumed to have desirable genes.
Exons portion of a gene included in the transcript of a gene and survives processing of the RNA in the cell nucleus to become part of a spliced messenger of astructural RNA in thecell cytoplasm; an exon specifies the amino acid sequence of a portion of the complete polypeptide. Fetal alcohol syndrome a link between excessive alcohol consumption during pregnancy and birth defects; characteristics include small head andeyes,foldsoftheskinthatobscuretheinner junctureoftheeyelids,short,upturnednose,and thin lips.
FISH florescent in situ hybridization: a techniqueforuniquelyidentifyingwholechromosomes or parts ofchromosomes using florescent tagged DNA.
Fragile sites
widththatusuallyinvolvesbothchromatidsandis always at exactly the same point on a specific chromosome derived from an individual or kindred.
Fragile-X syndrome mostcommonidentifiablecauseofgeneticmental deficiency.
Gamete an haploid cell.gel electrophoresis the process by which nucleic acids (DNA or RNA) or proteinsareseparatedbysizeaccordingtomovement of the charged molecules in an electrical field.
Gene a hereditary unit that occupies a certain position on a chromosome; a unit that has one or more specific effects on the phenotype, and can mutate to various allelic forms. Gene amplification any process by which specific DNA sequences are replicated disproportionatelygreaterthantheirrepresentationinthe parent molecules; during development, some genes become amplified in specific tissues.
Gene map the linear arrangement of mutable sites on a chromosome as deduced from genetic recombination experiments.
Gene therapy addition of a functional gene or group of genes to a cell by gene insertion to correct an hereditary disease.
Genetic counseling the educational process that helps individuals, couples, or families to understandgeneticinformationandissuesthatmay
a chromosome map showrelative positions of the known genes on
testinggroupsofindividuals toidentifydefectivegenescapableofcausinghea phenotypic variance of a trait in a population attributed to genetic heter-
all of the genes carried by a single gamete; the DNA content of an individual, which includes all 44 autosomes, 2 sex chromosomes,
genetic constitution of an organism. a sexcell or gamete (egg or spermatozoan).Haldane equation Haldane's law: the generalization that if first generation hybrids are producedbetweentwospecies,butonesexisabsent,rare,orsterile,thatsexistheheterogamic
the concept that both gene frequencies and genotype frequencies will remain constant from generation togeneration in an infinitely large, interbreeding population in which mating is at random and there is no selection, migration or mutation.
Hemophilia -- a sex-linked disease in humans in which theblood-clotting process is defective.

Unit
Heterogeneity the production of identical or similar phenotypes by different genetic mechanisms.
Heterozygote having two alleles that are different for a given gene.

HGP Human Genome Project. Homologous chromosomes chromosomes that pair during meiosis; each homologue isaduplicate of one chromosome from each parent.
more loci in homologous chromosome segments. Housekeeping genes all cells because they provide functions needed for sustenance of all cell types.
HUGO Human Genome Organization. Huntington disease irregular, spasmodic involuntary movements of thelimbsandfacialmuscles,mentaldeterioration anddeath,usuallywithin20yearsoftheonsetof symptoms.
Hybridization
labeledprobe(usuallyDNA)toitscomplementary sequence.
Ichthyosis
genital skin conditions; skin of affected individuals has a dry, scaly appearance. Imprinting allele which can be used to identify maternal or paternal origin of chromosome. In situ hybridization probe to its complementary sequence within intact, banded chromosomes.
Incomplete penetrance -- the gene for a condition is present, but not obviously expressed in all individuals in a familywith the gene.
Introns a segment of DNA (between exons) that is transcribed into nuclear RNA, but are removed in thesubsequent processing into mRNA.
Isochromosome a metacentric chromosome produced during mitosis or meiosiswhen the centromere splits transversely instead of longitudinally; the arms of such chromosome are equal in length and genetically identical, however,the loci are positioned in reverse sequence in the two arms.
Karyotype asetofphotographed,bandedchromosomesarrangedinorderfromlargesttosmall-
an endocrine condition
an extra X-chromosome (47,XXY); characterizedbythelackofnormalsexualdevelopment and testosterone, leading to infertility and adjustment problems if not detected and
analysis of pedigree the tracking of a roughafamilybyfollowingtheinheritance of a (closely associated) gene or trait and a DNA
the greater association in inheritance of two or more nonallelic genes than is to be expected from independent assortment; genes are linked because they reside on the same chromoanenzymethatfunctionsinDNArepair. logarithmoftheoddscore;ameasure of the likelihood of two loci being within a
autosomal dominant condition of connective tissue; affects the skeletal,
Marker-- agenewithaknownlocation on a chromosomeandaclear-cutphenotype,usedasapoint of reference when mapping a new mutant. Meiosis the doubling of gametic chromosome number.
Methylation addition of a methyl group (-CH3) to DNA or RNA.
Methylmalonic acidemia a group of conditions characterized by the inability to metabolize methylmalonic acid or by a defect in the metabolism of Vitamin B12.

Missense mutation a change in the base sequence of a gene that alters or eliminates a protein.
Mitochondrial DNA the mitochondrial genome consists of a circular DNA duplex, with 5 to 10 copies per organelle. Mitosis nuclear division. mRNA messenger RNA; anRNA molecular that functions during t quence of amino acids in a nascent polypeptide.
Multifactorial expressionbymanyfactors,bothgeneticandenvironmental.
Mutation -structural change. Myotonic dystrophy sive weakening or rigidity, with difficulty relaxing a contracted muscle; inherited Neurofibromatosis single gene condit oussystem;inmostcases,"cafeaulait"spots,are theonlysymptom;inheritedasanautosomaldominant trait,with 50%being new mutations. NIH National Institutes of Health. Nonsense mutation is changed to a stop codon, resulting in a truncated protein product. Noonan syndrome--aconditioncharacterizedby short stature and ovarian or testicular dysfunction, mental deficiency, and lesions of theheart. Northern analysis atechniquefortransferring electrophoretically resolved RNA segments from an agarose gel to a nitrocellulose filter paper sheet via capillary action.
Nucleotide one of the monomeric units from which DNA or RNA polymers are constructed; consists of apurine orpyrimidinebase, apentose sugar and a phosphoric acid group. Oncogenes genes involved in cell cycle control (growth factors, growth factor regulator genes, etc), a mutation can lead to tumor growth. Osteogenesis imperfecta a condition also knownasbrittlebonedisease;characterizedbya triangular shaped face with yellowish brown teeth, short stature and stunted growth, scoliosis, high pitched voice, excessive sweating and
thedevelopmentofanindividpolymerasechainreaction;atechniquefor copying the complementary strands of a target DNA molecule simultaneously for a series of cycles until thedesired amount is obtained. adiagramoftheheredityofaparticular trait through many generations of a family. observablecharacteristicsofanorganism produced by the organism's genotype in-
map where the distance between markers is the actual distance, such as the num-
phenylketonuria, an enzyme deficiency erized by the inability to convertoneaminoacid,phenylalanine,toanother,tyrosine, resulting in mental deficiency. plasmid double-stranded, circular, bacterial DNA into which afragment of DNA from another organism can be inserted.
Pleiotropy the phenomenon of variable phenotypes for a number of distinct and seemingly unrelated phenotypic effects.
Polycystic kidney disease (PKD) agroupofconditions characterized by fluid filled sacs that
Unit 8
slowly develop in both kidneys, eventually resulting in kidney malfunction.
Polymerase anyenzymethatcatalyzestheformation of DNA or RNA from deoxyribonucleotides or ribonucleotides.
Prader-Willi syndrome a condition characterized by obesity and insatiable appetite, mental deficiency, small genitals, andshortstature.May be deletion of#15 chromosome.
towards something in advance. Presymptomatic diagnosis netic condition toms.
Primer nucleotides used in the polymerase chainreactiontoinitiateDNAsynthesisataparticular location.
Probability
event relative to all alternative events, and usually expressed as decimal fraction. Proband -- individual in afamilywhobroughtthe family to medical attention. Probe single-stranded DNA labeledwithradioactive isotopes or tagged in other ways for ease in identification.

ble outcome of a disease.
Proteus syndrome distortedasymmetricgrowthofthebodyandenlarged head, enlarged feet, multiple nevi on the skin; mode of inheritance is unknown. Public policy that resultfrom theactions or lack of actionsof governmental entities.
Recessive a gene that is phenotypically manifestinthehomozygousstatebutismaskedinthe presence of a dominant allele.
Recombination thenaturalprocessofbreaking andrejoiningDNAstrandstoproducenewcombinations of genes and, thus, generate genetic variation. Gene crossover during meiosis. Repeat sequences the length of a nucleotide sequence that is repeated in a tandem cluster. Retinitis pigmentosa group of hereditary ocular disorders with progressive retinal degeneration. Autosomal dominant, autosomal recessive,
achildhoodmalignantcancerof the retina of the eye. reverse transcriptase viral
restriction fragment length polymorphism;variationsoccurringwithinaspeciesinthe length of DNA fragments generated by a species
oneoftheribonucleoprotein particles that are thesites of translation. condition with mull anomalies including: mental deficiency, broad thumbs, small head, broad nasal
"plus and minus" or "primed thod; DNA is synthesized so it is radioactivelylabeledandthereactionterminates specifically at the position corresponding to a
theprocess of determining therelative share allotted individuals of different genotypes in the propagation of a population; the selective effect of a gene can be defined by the probability that carriers of the gene will reproduce.
Sex determination the mechanism in a given species by which sex is determined; in many species sex is determined at fertilization by the nature of thesperm that fertilizes the egg. Sickle cell anemia an hereditary, chronic form of hemolytic anemia characterized by breakdown
Unit 8
of the red blood cells; red blood cells undergo a reversible alteration in shape when the oxygen tension of the plasma falls slightly and a sicklelike shape forms.
Somatic cell hybrid hybrid cell line derived from two different species; contains a complete chromosomal complement of one species and a partial chromosomal complement of the other; human/hamster hybrids grow and divide, losing human chromosomes with each generation until they finally stabilize, the hybrid cell line established is then utilized to detect the presence of genes on there
Somatic mutation cell that is not destined to become a germ cell;if the mutant cell continues to divide, theindividual willcometocontainapatchoftissueofgenotype different from the cells of therest of thebody. Southern blotting electrophore an agarose gel to a nitrocellulose filter paper sheetviacapillaryaction;theDNAsegmentofinterest is probed with a radioactive, complementary nucleic acid, and its position is determined by autoradiography.
Spina bifida from altered fetal development of the spinal cord, part of the neural plate fails to join together and bone and muscle are unable to grow over this open section.
Syndrome multiple signs, sympt characterize a particular condition; syndromes arethoughttoarisefromacommonoriginandresultfrommorethanonedevelopmentalerrorduring fetal growth.
Tay-Sachs disease -- a fatal degenerative diseaseofthenervoussystemduetoadeficiencyof hexosamidaseA,causingmentaldeficiency,paralysis, mental deterioration, and blindness; found
primarily but not exclusively among Ashkenazi Jews.Autosomal recessive.
Teratogens anyagentthatraisestheincidence of congenital malformations.
Trait anydetectablephenotypicpropertyofan organism.
Transcription the formation of an RNA moleculeuponaDNAtemplatebycomplementarybase
ransfer ofbacterialgenetic material from one bacterium to another using a
enzymes thatcatalyze thetransfer of functional groups between donor and ac-
one into which a cloned genetic material has been experimentally transferred, a subset of these foreign gene express themselves intheir offspring. Turner syndrome a chromosomalconditioninfemales(usually45,XO) due to monosomy of the X- chromosome; characterized by short stature, failure to develop secondary sexcharacteristics, and infertility. the formation of a polypeptide ificaminoacidsequencedirected by the genetic information carried by mRNA. a chromosome aberration which results in a change in position of a chromosomal segment within the genome, but does not change
a code in which a given amino acid setof three nucleotides. Tumor suppressor gene -- genes that normally function to restrain the growth of tumors; the best understood case is for hereditary retinoblastoma.
Vector -- a self-replicating DNA molecule that transfers a DNA segment between host cells.

VNTR variable number tandem repeats; any gene whose alleles contain different numbers of tandemly repeated oligonucleotide sequences. Von Hippel-Lindau syndrome an autosomal dominant condition characterized by the anomalous growth and proliferation of blood vessels on the retina of the eye and the cerebellum of the brain;cystsandcancersinthekidneys,pancreas, and adrenal glands.
Western blotting analysis identify a specific pr actively labeled antibody raised against the protein in question.
X-inactivation X-chromosomesint amethodofdosagecompensation;atanearlyembryonicstageinthenormalfemale,oneofthetwo X-chromosomes undergoes inactivation, apparentlyatrandom,fromthispointonalldescendent cells will have the same X-chromosome inactivated as the cell from which they arose, thus a femaleisamosaiccomposedoftwotypesofcells, onewhichexpressesonly thepaternalX-chromosome, and another which expresses only the maternal X-chromosome.
XYY syndrome extra Y chromosome (in 1 in 1000 male births).
Symptoms: tall stature (over 6'), may including sterility,developmental delay, learning problems. YAC yeast artificial chromosome; a linear vector into which a large fragment of DNA can be inserted; the development of YAC's in 1987 has increasedthenumberofnucleotideswhichcanbe cloned.
Zoo blot northern analysis of mRNA from dif-

