AlgebraicmorphologyofDNA–RNAtranscriptionand regulation
MichelPlanat 1,*,† ,MarceloM.Amaral 2,† andKleeIrwin 2

1 InstitutFEMTO-STCNRSUMR6174,UniversitédeBourgogne-Franche-Comté,F-25044Besançon,France
2 QuantumGravityResearch,LosAngeles,CA90290,USA;marcelo@quantumgravityresearch.org(M.M.A.); klee@quantumgravityresearch.org(K.I.)

* Correspondence:michel.planat@femto-st.fr
†Theseauthorscontributedequallytothiswork.
Abstract: Transcriptionfactors(TFs)andmicroRNAs(miRNAs)areco-actorsingenome-scale 1 decodingandregulatorynetworks,oftentargetingcommongenes.Inthispaper,wedescribe 2 thealgebraicgeometryofbothTFsandmiRNAsthankstogrouptheory.InTFs,thegeneratorof 3 thegroupisaDNA-bindingdomainwhile,inmiRNAs,thegeneratoristheseedofthesequence. 4 Forsuchagenerated(infinite)group π,wecomputethe SL(2, C) charactervariety,where SL(2, C) 5 issimultaneouslya‘space-time’(aLorentzgroup)anda‘quantum’(aspin)group.Anoteworthy 6 resultofourapproachistorecognizethatoptimalregulationoccurswhen π lookslikeafreegroup 7 Fr (r = 1 to 3)inthecardinalitysequenceofitssubgroups,aresultobtainedinourpreviouspapers. 8 Anonfreegroupstructurefeaturesapotentialdisease.Asecondnoteworthyresultisaboutthe 9 structureoftheGroebnerbasis G ofthevariety.Asurfacewithsimplesingularities(likethewell 10 knownCayleycubic)within G isasignatureofapotentialdiseaseevenwhen G lookslikeafree 11 group Fr initsstructureofsubgroups.Ourmethodsapplytogroupswithageneratingsequence 12 madeoftwotofourdistinctDNA/RNAbasesin {A, T/U, G, C}.SeveralhumanTFsandmiRNAs 13 areinvestigatedindetailthankstoourapproach. 14
Keywords: Transcriptionfactors;micro-RNAs;diseases;finitelygeneratedgroup; SL(2, C) character 15 variety,algebraicsurfaces 16
Citation: Planat,M.;Amaral,M.M.; Irwin,K.Algebraicmorphologyof DNA–RNAtranscriptionand regulation. Int.J.Mol.Sci. 2022, 1,0. https://doi.org/
Received: Accepted: Published:
Publisher’sNote: MDPIstaysneutral withregardtojurisdictionalclaimsin publishedmapsandinstitutionalaffiliations.
Copyright: © 2022bytheauthors. Submittedto Int.J.Mol.Sci. for possibleopenaccesspublication underthetermsandconditions oftheCreativeCommonsAttribution(CCBY)license(https:// creativecommons.org/licenses/by/ 4.0/).
0.Introduction
17
Recently,wewroteapaperaboutacommonalgebrapossiblyrulingthebeautyand 18 structureinpoems,musicandproteins[1].Wefoundthatfreegroupsgovernthestructure 19 ofsuchdisparatetopicswherealanguageemergesfrompurerandomness.Wecoined 20 theconceptof‘syntacticalfreedom’forqualifyingthisoccurrenceofsymbolsorganized 21 accordingtoaperiodicity[2,3].Accordingtoourview,theescapeto‘syntacticalfreedom’ 22 meansalackofbeautyandthesignatureofapotentialdiseaseatthegenomescale.The 23 secondarystructuresinthesequenceofproteinsorvirusesare,mostofthetime,organized 24 accordingtotherulesoffreegroupsand,otherwisetheymaybeawitnessofapotential 25 aberranttopology.Onefavoritedecompositionofthesecondarystructureofproteinsisin 26 termof α-helices, β-sheetsandcoils[4]butthisdecompositionandtheresultingsyntaxis 27 modeldependent[1]. 28
ApartfromthecanonicaldoublehelixB-DNAwenowknowthatthereexistsa 29 diversityofnon-canonicalcoding/decodingsequencesorganizedinstructuressuchas 30 Z-DNA(oftenencounteredintranscriptionfactors),G-quadruplex(intelomeres)andother 31 typesthataresingle-stranded,two-strandedormultistranded[5].RNAisusuallyasingle- 32 strandedmoleculeinashortchainofnucleotides,asisthecaseforamessengerRNA 33 (mRNA)ora(non-coding)microRNA(miRNA). 34
Inourapproach,theinvestigatedsequencesdefineafinitelygeneratedgroup f p 35 whosestructureofsubgroupsiscloseorawayfromafreegroup Fr ofrank r,where r + 1 is 36
VersionDecember13,2022submittedto
https://www.mdpi.com/journal/ijms
© 2022 by the
Distributed under a Creative Commons CC BY
Int.J.Mol.Sci.
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
author(s).
license.
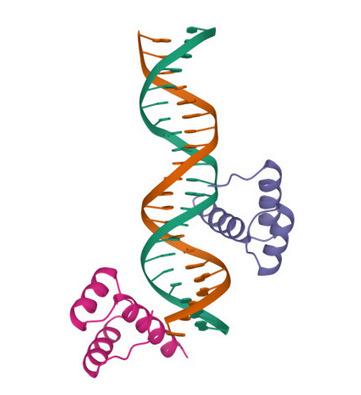
Figure1. Left:theNanogtranscriptionfactor(PDB9ANT).Right:thepre-miR-155secondary structure[10].
thenumberofdistinctaminoacidsinthesequence(orthenumberofdistinctsecondary 37 structuresconsideredintheproteinchain).Recently,wealsointroducedconceptsfor 38 representingthegroups f p overtheLiegroup SL(2, C).The SL(2, C) charactervariety 39 of f p anditsGroebnerbasisaretopologicalingredients,theyfeaturealgebraicgeometric 40 propertiesofthegroup f p underquestion[6,7].ForthedefinitionoftheGroebnerbasisof 41 anidealcontainingmultivariatepolynomialrings,thereadermayconsultReference[8]. 42 Inthispaper,wewillfocusupontranscriptionfactorsandmiRNAs,bothserveat 43 properlydecodingandregulatingthegenesandtheiraction,eitherindependentlyof 44 eachotherortogetherbytargetingcommongenes[9].Figure 1 (Left)isapictureofthe 45 pluripotenttranscriptionfactorNanog.Figure 1 (Right)isanexampleofapre-miRNA 46 associatedtoadisease[10].Bothwillbeinvestigatedindetailinthispaper. 47

InSection 1,wedescribethemathematicalmethodsandthesoftwareneededfor 48 describingthealgebraicsurfacesrelevanttoDNA/RNAsequences.Thisincludesthe 49 definitionofinfinitegroupsunderquestion,ofthefreegroups Fr ofrank r = 1 to 3 50 correspondingto 2-to 4-basesequencesandthecalculationof SL(2, C) representationof 51 suchgroups.AspecialcareisneededtocomputeaGroebnerbasisofthecharactervariety. 52
Section 2 isadiscussionaboutthetypeofsingularsurfacesencounteredinthisresearch. 53 Theyplayanimprtantroleinourviewofapotentialdisease. 54
InSection 3,themethodsareappliedtorepresentativeexamplesofsequencestaken 55 fromtranscriptionfactorsandmicroRNAswhosegroupiscloseorawayfromafreegroup, 56 andwhoseGroebnerbasisofthevarietycontainssimplesingularities.Figures 2 to 6 feature 57 themaintopologicalingredientsresultingfromthisresearch. 58
VersionDecember13,2022submittedto Int.J.Mol.Sci. 2of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
1.MaterialsandMethods
59
1.1.Finitelygeneratedgroups,freegroupsandtheirconjugacyclasses 60
Afreegroup Fr on r generators(ofrank r)consistsofalldistinctwordsthatcanbe 61 builtfrom r letterswheretwowordsaredifferentunlesstheirequalityfollowsfromthe 62 groupaxioms.Thenumberofconjugacyclassesof Fr ofagivenindex d isknownandis 63 agoodsignatureoftheisomorphism,orthecloseness,ofagroup π to Fr [3,11].Inthe 64 following,thecardinalitystructureofconjugacyclassesofindex d in Fr iscalledthecard 65 seqof Fr,andweneedthecasesfrom r = 1 to 3 tocorrespondtothenumberofdistinct 66 basesinaDNA/RNAsequence.Thecardseqof Fr isinTable 1 forthe 3 sequencesof 67 interestinthecontextofDNA/RNA. 68
Table1. Numberofconjugacyclassesofsubgroupsofindex d infreegroupofrank r = 1 to 3.The lastcolumnistheindexofthesequenceintheon-lineencyclopediaofintegersequences[12].
r cardseq sequencecode
1 [1,1,1,1,1,1,1,1,1, ] A000012
2 [1,3,7,26,97,624,4163,34470,314493, ] A057005 3 [1,7,41,604,13753,504243,24824785,1598346352, ··· ] A057006
Next,givenafinitelygeneratedgroup fp witharelation(rel)givenbythesequence 69 motif,weareinterestedinthecardinalitysequence(cardseq)ofitsconjugacyclasses. 70 Often,theDNA/RNAmotifinthesequenceunderinvestigationisclosetothatofa 71 freegroup Fr,with r + 1 beingthenumberofdistinctbasesinvolvedinthemotif.But 72 thefinitelygeneratedgroup f p = ⟨x1, x2|rel(x1, x2)⟩,or f p = ⟨x1, x2, x3|rel(x1, x2, x3)⟩,or 73 f p = ⟨x1, x2, x3, x4|rel(x1, x2, x3, x4)⟩ (wherethe xi aretakeninthefourbasesA,T/U,G,C 74 andrelisthemotif)maynotbethefreegroup F1 = ⟨x1, x2|x1 x2⟩,or F2 = ⟨x1, x2, x3|x1 x2 x3⟩, 75 or F3 = ⟨x1, x2, x3, x4|x1 x2 x3 x4⟩.Theclosenessof f p to Fr canbecheckedbyitssignaturein 76 thefiniterangeofindicesofthecardseq. 77
1.2.TheSL(2, C) charactervarietyofafinitelygeneratedgroupandaGroebnerbasis 78
Let f p beafinitelygeneratedgroup,wedescribetherepresentationsof f p inthe 79 (doublecoverofthe)Lorentzgroup SL(2, C),thegroupof (2 × 2) matriceswithcomplex 80 entriesanddeterminant 1.Thegroup SL(2, C) maybeseensimultaneouslyasa‘space-time’ 81 (aLorentzgroup)anda‘quantum’(aspin)group. 82
Suchagroupcontainsrepresentationsasdegreesoffreedomforallquantumfields 83 andisthegaugegroupforEinstein-CartantheorywhichcontainstheEinstein-Hilbert 84 actionandEinstein’sfieldequations[13].TheHolstactionusedinloopquantumgravity 85 hasquantumgravitystatesgivenintermsof SL(2, C) representations[14]. 86
Representationsof f p in SL(2, C) arehomomorphisms ρ : f p → SL(2, C) withchar- 87 acter κρ (g)= tr(ρ(g)), g ∈ f p.Thesetofcharactersallowstodefineanalgebraicsetby 88 takingthequotientofthesetofrepresentations ρ bythegroup SL(2, C),whichactsby 89 conjugationonrepresentations[15,16]. 90
Fortheeffectivecalculationsofthecharactervariety,wemakeuseofasoftwareon 91 Sage[17].WealsoneedMagma[18]forthecalculationofaGroebnerbasis,atleastfor 3- 92 and4-basesequences. 93
Following[19],inthissection,wedescribethespecialcaseofrepresentationsforthe 96
1,1)
Letustaketheexampleofthepuncturedtorus T1,1 whosefundamentalgroup,that wedenote π,isthefreegroup F2 = ⟨a, b|∅⟩ ontwogenerators a and b.Theboundary
VersionDecember13,2022submittedto Int.J.Mol.Sci. 3of13
1.3.AlgebraicgeometryandtopologyofDNA/RNAsequences 94 1.3.1.Two-basesequences 95
puncturedtorus
S1,1 andtherelevanceoftheextendedmappingclassgroup Mod±(S
97 initsactiononsurfacesoftype κd (x, y, z), d ∈ C. 98
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
componentof T1,1 isasinglelooparoundthepunctureexpressedbythecommutator [a, b]= abAB with A = a 1 and B = b 1.Weintroducethetraces
x = tr(ρ(a)), y = tr(ρ(b)), z = tr(ρ(ab)).
Thetraceofthecommutatoristhesurface[15,19]
tr([a, b])= κ2(x, y, z)= x2 + y2 + z2 xyz 2.
Anothernoticeablesurfaceisobtainedfromthecharactervarietyattachedtothe fundamentalgroupoftheHopflink L2a1 thatlinkstwounknottedcurves.FortheHopf link,thefundamentalgroupis
π(S3 \ L2a1)= ⟨a, b|[a, b]⟩ = Z2 , andthecorrespondingcharactervarietyistheCayleycubic[6] κ4(x, y, z)= x2 + y2 + z2 xyz 4.
Surfaces κ2 and κ4 havebeenobtainedfromtwodifferentmathematicalconcepts,from topologicalandalgebraicconceptsindimension 2,respectively.Torelatethemonemakes useoftheDehn-Nielsen-Baertheoremappliedtotheoncepuncturedtorus[20].According tothistheorem,forasurfaceofgenus g ≥ 1,wehave
Mod±(Sg ) ∼ = Out(π(Sg )),
wherethemappingclassgroup Mod(S) denotesthegroupofisotopyclassesoforientation- 99 preservingdiffeomorphismsof S (thatrestricttotheidentityontheboundary ∂S if ∂S = ∅), 100 theextendedmappingclassgroup Mod±(S) denotesthegroupofisotopyclassesofall 101 homeomorphismsof S (includingtheorientation-reversingones)and Out(π) Thisleadsto 102 the(topological)actionofMod± onthepuncturedtorusasfollows 103
Mod±(S1,1)= Out(F2)= GL(2, Z).(1)
Theautomorphismgroup Aut(F2) actsbycompositionontherepresentations ρ andinducesanactionoftheextendedmappingclassgroup Mod± onthecharactervarietyby polynomialdiffeomorphismsofthesurface κd definedby[19] f (4) H (x, y, z)= κd (x, y, z)= xyz x2 y2 z2 + d.(2)
TheCayleysurface κ4(x, y, z) possesses 4 simplesingularities.Wealreadyshowed 104 thatitplaysaroleinthecontextofZ-DNAconformationsoftranscriptionfactors[7,Tables 105 2and5].Seealsothesection 3 belowandnotablyFigures 3 (Left)and 6 (Left). 106
Thesurface κ3(x, y, z) lieswithinthecharactervarietyforthefundamentalgroupof 107 thelinkL6a1[21].WeshowbelowthatthissurfacealsoliesinthegenericGroebnerbasis 108 obtainedfor4-basesequences,seeFigure 6 (Right)below. 109
1.3.2.Three-basesequences
110
Ourmainobjectinthissectionisthefourpuncturedsphereforwhichthefundamental 111 groupisthefreegroup F3 ofrank 3 whosecharactervarietygeneralizestheFrickecubic 112 surface(2)tothehypersurface Va,b,c,d (C) in C7 113 Wefollowtheworkofreferences[15,19,22]. 114
Let S4,2 bethequadruply-puncturedsphere.Thefundamentalgroupfor S4,2 canbeex- 115 pressedintermsoftheboundarycomponents A, B, C, D as π(S4,2)= ⟨A, B, C, D|ABCD⟩ ∼ = 116 F3. 117
VersionDecember13,2022submittedto Int.J.Mol.Sci. 4of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
Arepresentation π → SL(2, C) isaquadruple
α = ρ(A), β = ρ(B) γ = ρ(C), δ = ρ(D) ∈ SL(2, C) where αβγδ = I
Letusassociatetheseventraces 118
a = tr(ρ(α)), b = tr(ρ(β)), c = tr(ρ(γ)), d = tr(ρ(δ)) x = tr(ρ(αβ)), y = tr(ρ(βγ)), z = tr(ρ(γα)),
where a, b, c, d areboundarytracesand x, y, z aretracesofelements AB, BC, CA represent- 119 ingsimpleloopson S4,2. 120 Thecharactervarietyfor S4,2 satisfiestheequation[15,Section5.2],[19,Section2.1],[22, Section3B],[23,Eq.1.9]or[24,Eq.(39)]
Va,b,c,d (x, y, z)= x2 + y2 + z2 + xyz θ1 x θ2y θ3z θ4 = 0(3) with θ1 = ab + cd, θ2 = ad + bc, θ3 = ac + bd and θ4 = 4 a2 b2 c2 d2 abcd 121 The 4-puncturedsphere,whosefundamentalgroupisthefreegroup F3 withgenerator 122 theproductofthe 4 letters,isagenerictopology.Itisstraightforwardtocheckthatthe 123 Groebnerbasisfor F3 contains(amongothersurfacesanddependingonthechoiceof 124 parameters)asinglecopyofthegenericsurfaces κ4(x, y, z), κ3(x, y, z) and V1,1,1,1(x, y, z)= 125 xyz + x2 + y2 + z2 2x 2y 2z + 1,asurfacewealsodenote f (3A1) (x, y, z) becauseit 126 contains3simplesingularitiesoftype A1 asshowninFigures 2 and 6 (Right). 127
ThereareothersurfacesencounteredinourstudyoftheGroebnerbasisfortranscrip- 128 tionfactorsandmiRNAswhenthegeneratedgroupiscloseorawayfromthefreegroup 129 F2 (for3-basesequences)orthefreegroup F3 (for 4 basesequences).Thesesurfacesare 130 describedinSection 3 131
1.3.3.Four-basesequences
132
TheredoesnotexistahugedifferenceinthestructureofaGroebnerbasisofthe 133 charactervarietyinthecaseofa 4-basesequencecomparedtothecaseofa 3-basis 134 sequence.Onedifferenceisthatonehastomanagea 14-dimensionalhypersurface 135 Va,b,c,d,e, f ,g,h (x, y, z, u, v, w) in C14 (insteadofa 7-dimensionaloneasintheprevioussub- 136 section).Ingeneral,aftertheappropriatechoiceofthe 8 parameters a, b, c, d, e, f , g, h,the 137 GroebnerbasiscontainsmorethanonecopyofthegenericGroebnerbasis,asshownin 138 Table 4.Eachcopy S ofarelevantsurfacemaybeoftheform S(x, y, z), S(x, u, v), S(y, u, w) 139 or S(z, v, w) 140
2.Discussion
141
Givenanordinaryprojectivesurface S intheprojectivespace P3 overanumberfield, 142 if S isbirationallyequivalenttoarationalsurface,thesoftwareMagma[18]determinesthe 143 maptosucharationalsurfaceandreturnsitstypewithinfivecategories.Thereturned 144 typeof S is P2 fortheprojectiveplane,aquadricsurface(foradegree 2 surfacein P3),a 145 rationalruledsurface,aconicbundleoradegree p DelPezzosurfacewhere1 ≤ p ≤ 9. 146
Afurtherclassificationmaybeobtainedfor S in P3 if S hasatmostpointsingularities. 147 Magmacomputesthetypeof S (orrather,thetypeofthenon-singularprojectivesurfaces 148 initsbirationalequivalenceclass)accordingtotheclassificationofKodairaandEnriques 149 [25].ThefirstreturnedvalueistheKodairadimensionof S,whichis ∞, 0, 1 or 2.The 150 secondreturnedvaluefurtherspecifiesthetypewithintheKodairadimension ∞ or 0 151 cases(andisirrelevantintheothertwocases). 152
Kodairadimension ∞ correspondstobirationallyruledsurfaces.Thesecondreturn 153 inthiscaseistheirregularity q ≥ 0 of S.So S isbirationallyequivalenttoaruledsurface 154 overasmoothcurveofgenus q andisarationalsurfaceifandonlyif q iszero. 155
VersionDecember13,2022submittedto Int.J.Mol.Sci. 5of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
Kodairadimension 0 correspondstosurfaceswhicharebirationallyequivalenttoa K3 156 surface,anEnriquessurface,atorusorabi-ellipticsurface. 157
EverysurfaceofKodairadimension 1 isanellipticsurface(oraquasi-ellipticsurface 158 incharacteristics 2 or 3),buttheconverseisnottrue:anellipticsurfacecanhaveKodaira 159 dimension ∞,0or1. 160
SurfacesofKodairadimension2arealgebraicsurfacesofgeneraltype. 161
Oneimportantattributeofasurfaceisitsdegreeofsingularity.Mostsurfaces S 162 ofinterestbelowarealmostnotsingularinthesensethattheyhaveatworstsimple 163 singularities.Thetypeandthenumberofsimplesingularitiesaredenotedinanexponent 164 suchas S(lA1) forlsingularitiesoftype A1.Thenotation A1 referstothetypeofaCoxeter 165 rootsystem.Noothertypesofsingularsurfacesareencounteredinourpaper.Allsuch 166 surfacearedegree 3 delPezzosurfaces.FortheCayleycubic f (4) (x, y, z),theexponent (4) 167 means (4A1).Ofcoursetherearenonsingularsurfacesinthecharactervarietyforsome 168 sequences,correspondingtotheothertypes.Butwedonotdiscusstheminthispaper. 169
ThereareadditionalfacilitiesofferedbyMagmaforstudyingasingularscheme.A 170 schemeinMagmaisanygeometricobjectdefinedbythevanishingofpolynomialsin 171 aprojectivespace.Analgebraicsurfaceisascheme.Iftheschemeissingular,onecan 172 calculateitssingularsubscheme,itsdegreeanditssupport,thatareimportantsignatures 173 ofthescheme.Mostofthetime,intheexamplesofthispaper,forasingularsurface S(lA1) , 174 thedegreeis l andthesupportcontains l simplesingularpoints.Otherwise,weadda 175 lowerindexto S(lA1) toqualifytheindexandthesupportofthesingularsubscheme.The 176 notation S(lA1) m,{} meansthatthereare l singularitiesoftype A1,thatthedegreeofthesingular 177 subschemeis m andthatthesupportistheemptyset. 178
Bytheway,transcriptionfactorsandmicroRNAsseemtohavesmoothrulesunderlyingthesingularitiesoftheirsequences.Thiscontrastswithsomesequencesencounteredin anothercontext.In[7],wefoundtwocasesoftwo-basesequenceswhosecorresponding Groebnerbasiscontainsthesurfaces
f H (x, y, z)= z4 2xyz (+z3)+ 2x2 + 2y2 3z2( 4z) 4.(4)
Aplotofoneofthemin[7,Figure3(Right)]showsthattheylooklikeageneralization 179 f H (x, y, z) oftheCayleycubic f (4) H (x, y, z).Thesesurfacesareconicbundlesinthefamilyof 180 K3 surfaces.Incontrastto f (4) H (x, y, z),accordingtoMagma,theresolutionofsuchsingular 181 surfaces f H (x, y, z) leadstomanynonisolatedsingularities(onecannotdesingularisethe 182 surfacesbyblowup). 183
Tosummarizetheimportantissuesbelow,anoticeableresultofourapproachisto 184 recognizethatoptimalregulationoccurswhenthegroupunderlyingthesequencelooks 185 likeafreegroup Fr (r = 1 to 3)inthecardinalitysequenceofitssubgroups,aresultobtained 186 inourpreviouspapers.Anonfreegroupstructurefeaturesapotentialdisease.Asecond 187 noticeableresultisaboutthestructureoftheGroebnerbasisofthevariety.Asurfacewith 188 simplesingularities(likethewellknownCayleycubic)withintheGroebnerbasisisa 189 signatureofapotentialdiseaseevenwhenthegeneratedgrouplookslikeafreegroup Fr 190 initsstructureofsubgroups.Ourmethodsapplytogroupswithageneratingsequence 191 madeoftwotofourdistinctDNA/RNAbasesin {A, T/U, G, C}.SeveralhumanTFsand 192 miRNAsareinvestigatedindetailthankstoourapproach. 193
3.Results
194
Inthissection,weapplythe SL(2, C) representationtheorytogroupsgeneratedby 195 DNA/RNAsequencesoccurringintranscriptionfactorsandmicroRNAs.Bothplaya 196 leadingroleinthedecodingofthegenomeandingenome-scaleregulatorynetworks. 197 Two-lettertranscriptionfactors(TFs)whosestructureiscloseorawayfromthefreegroup 198 F1 werealreadyinvestigatedin[7,Table2].TheoccurrenceoftheCayleycubic κ4(x, y, z) 199 intheGroebnerbasisofthecharactervarietywasfoundtobeasignatureintheformer 200 case.Inthiscasethissurfaceseemstopossessaregulatoryactionthatmaybelostinthe 201
VersionDecember13,2022submittedto Int.J.Mol.Sci. 6of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
lattercase.In[7,Table3],thepotentialdiseasesassociatedtoanon-freegroupstructureare 202 mentioned.Inthepresentsection,oneexploresinTable 2 thecaseofa 3-lettersequenceof 203 aTFandinTable 3 thecaseof 3-lettersequenceofamiRNA.Thecaseof 4-lettersequence 204 ofamiRNAissummarizedinTable 4.Theroleofsurfaceswithsimplesingularitiesinthe 205 Groebnerbasisisemphasized. 206
3.1.AlgebraicmorphologyofthetranscriptionfactorPrdm1
207
ThetranscriptionalrepressorPRdomaincontaining1(Prdm1),alsoknownasB- 208 lymphocyte-inducedmaturationprotein-1(Blimp1),isessentialfornormaldevelopment 209 andimmunity[26].Itisofazincfingertype.TheconsensussequenceACTTTCcorresponds 210 tothecodeMA0508.2in[27]. 211
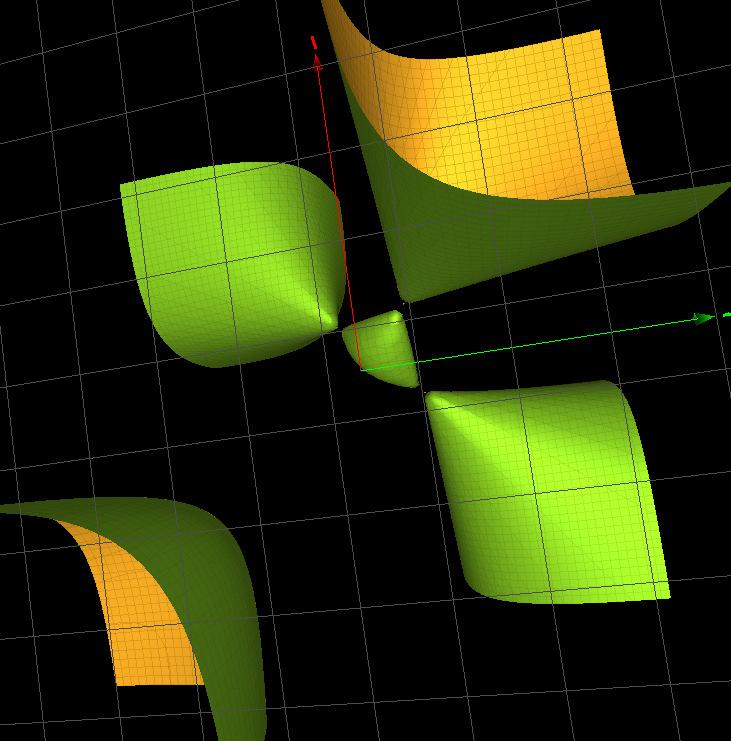
Figure2. TheFrickesurface V1,1,1,1(x, y, z)= f (3A1 ) a (x, y, z) (withthreesimplesingularitiesoftype A1).
Thecharactervariety
212
Theidealforthecharactervariety fPrdm1(a, b, c, d)(x, y, z) forafewvaluesofthe 213 parametersis 214
fPrdm1(0,0,0,0)= κ 4(x, y, z)(yz + x + 2), fPrdm1(0,1,1,0)= yκ 2(x, y, z)(x 1),
fPrdm1(0,1,0,0)= zκ 3(x, y, z)(z2 + 1)(yz + x + 1)(yz + x + 2), fPrdm1(1,1,1,1)= f (3A1) a (x, y, z)(y + 1)(y + z 1), where κ 2(x, y, z), κ 3(x, y, z) areFrickesurfaces[21]and f (3A1) a (x, y, z)= xyz + x2 + 215 y2 + z2 2x 2y 2z + 1 isthesurfacedrawninFigure 2.Thesubscript 3A1
VersionDecember13,2022submittedto Int.J.Mol.Sci. 7of13
218 ThesingularsurfacesfoundintheGroebnerbasisoftheidealarenotsimilartothose 219 intheideal.Oneofthem S1 = S(A1 A3) 3,
} = 2yz2 + x2 + 3z2 2xz 2yz 2
2z
220 obtainedatvalues
featuringtwosimplesingularitiesoftype
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
isforfeaturing 216 thethreesingularitiesoftype A1. 217 TheGroebnerbasis
{0:1:0:1
y
2 is
(a, b, c, d)=(1,1,1,1)
A1 and 221 A3 withasingularsubschemeofdegree 3 andthesingularpointoftype A3 initssupport. 222 TheothersurfaceobtainedintheGroebnerbasisatvalues (a, b, c, d)=(0,0,0,0) isaconic 223
bundleofthe K3 type S2 := z4 + 2yz3 + x2 6yz 2x 8 whosesingularsubschemeis 224 nonzerodimensionalandofdegree1. 225
ThesetwosurfacesarenonstandardinourcontextofTFsandmiRNAs. 226
3.2.AlgebraicmorphologyofhomeodomainsforNanogandXvent 227
Thepluripotencyinembryonicstemcellsandtheirregulationischaracterizedbythe 228 expressionofseveraltranscriptionfactors[28,29].Amongthem,thetranscriptionfactor 229 Nanogispresentintheembryonicstagesoflifeofseveralvertebratespecies.Nanogbinds 230 topromoterelementsofhundredsoftargetgenesasaregulatoryelement.Ithasaconserved 231 DNA-bindinghomeodomainwithconsensussequenceTAATGG.Theclosesthomolog 232 ofNanogisthe(nonmammalia)Xenopus,aXventtranscriptionfactorwithconsensus 233 sequenceCTAATT[29].Inthissubsection,weinvestigatethealgebraicmorphologyofboth 234 transcriptionfactorsNanogandXventthankstotheirconsensussequences. 235
Table2. Afew(three-base)transcriptionfactorswhosegroupstructureisawayfromafreegroup orwhoseGroebnerbasisofthe SL(2, C) charactervarietycontainsa(possiblyalmost)singular surface.ThesymbolgeneisfortheidentificationofthetranscriptionfactorintheJaspardatabase [27],motifisfortheconsensussequenceofthetranscriptionfactor,cardseqisforthecardinality sequenceofconjugacyclassesofsubgroupsofthegroupwhosemotifisthegenerator,singisforthe identificationofasingularsurfacewithintheGroebnerbasis,thelastcolumnnisforareferencepaper andthecorrespondingdisease.Thegroup F2 isthefreegroupofranktwo.Thecardseqfor π2 is [1, 3, 10, 51, 164,1230,7829,59835,491145 ],closetothecardseqofthegroup ⟨x, y, z|(x, (y, z))= z⟩. Thelatergroupisfoundasgoverningthestructureofmanytranscriptionfactorsandisassociatedto thelinkfoundin[7,Figure2].Thecardseqfor π3 is [7,14,89,264,1987,11086,93086 ].Thesurface f (A1 ) b (x, y, z)= x2 + y2 6z2 + 4xyz (notdefinedinthetext)ispartofthecharactervarietyforthe genesPitx1,OTX1,...
gene motif cardseq singref&disease Prdm1 ACTTTC F2 S1, S2(x, y, z) [27],MA0508.2 lupus,rheumatoidarthritis POU6F1 TAATGAG π2 nosing.,MA1549.1 lungadenocarcinoma ELK4 CTTCCGG . nosing,Fricke .,MA0076.2 gastriccancer OTX2 GGATTA π3 nosing .,[MA0712.2,MA0883.1] medulloblastomas N-box TTCCGG nosing,Fricke [30] drugsensitivity Pitx1,OTX1, TAATCC. f (4) H , f (A1 ) b (x, y, z) [27],[MA0682.1,MA0711.1] autism,epilepsy, Nanog TAATGG f (4) H , f (A1 ) a (x, y, z) [28] cancercells Xvent CTAATT F2 f (2A1 ) 4,{} , f (A2 ) (x, y, z) [29]
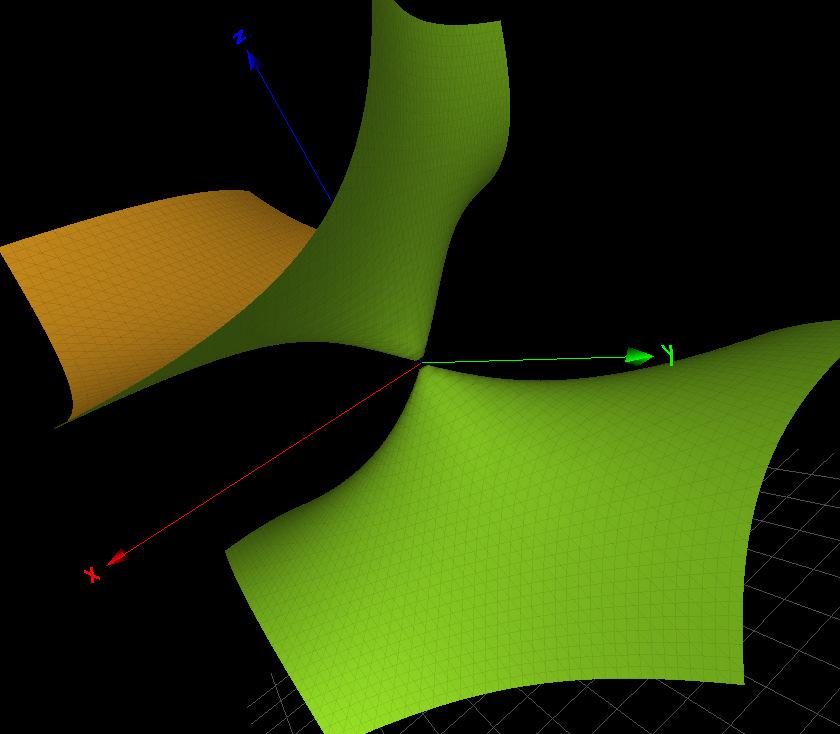
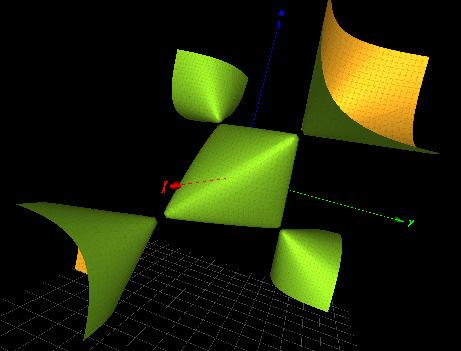
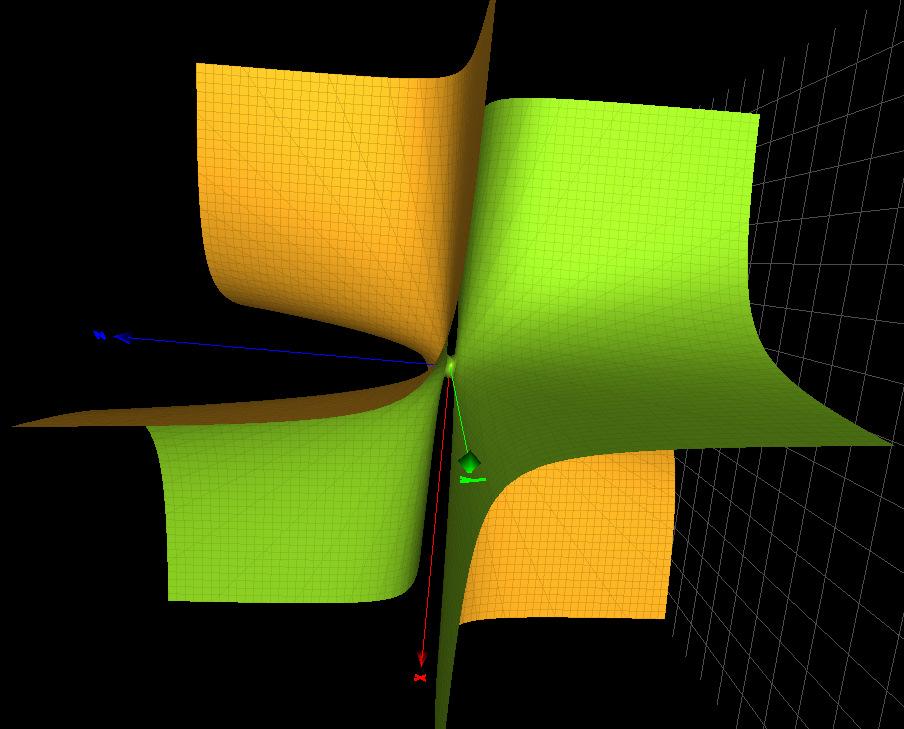
TheGroebnerbasisforXvent fNanog(0,0,0,0) takestheform 236 fNanog(0,0,0,0)= f (4) H (x, y, z) f (A1) a (x, y, z) where f (4) H (x, y, z) istheCayleycubic(withits 4 simplesingularities)and f (A1) a (x, y, z)= 237 x2 + y2 z2 + xyz (asurfacewithasinglesimplesingularityoftype A1)asshowninFigure 238 3 (Right).Theforgottenfactorsarefactorsforplanesortrivialsmoothsurfaces. 239 TheGroebnerbasisforXvent fXvent(1,1,1,1) takestheform 240 fXvent(1,1,1,1)= f (3A1) b (x, y, z) ···
VersionDecember13,2022submittedto Int.J.Mol.Sci. 8of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022
doi:10.20944/preprints202212.0256.v1
Figure3. Left:theCayleycubic κ4(x, y, z).Right:thesurface f (A1 ) a (x, y, z) where f (3A1) b (x, y, z)= x2 + y2 + xyz xy z 1 (asurfacewiththreesimplesingularity 241 oftype A1).Themissingtermdoesnotcontainsurfaceswithsingularities.Thecharacter 242 variety fXvent(0,0,0,0) containsthecubicsurface f (2A1) 4,{} (x, y, z)= 2z3 + x2z + 2xyz + 243 2y2 z2 6z (withtwosimplesingularitiesoftype A1)andotherfactorsforplanesor 244 trivialsmoothsurfaces.Bothsurfaces f (3A1) b (x, y, z) and f (2A1) 4,{} (x, y, z) arepicturedinFigure 245 4 246 Figure4. Left:thecubicsurface f (2A1 ) 4,{} (x, y, z).Right:thecubicsurface f (3A1 ) b (x, y, z) Table 2 listsafewselectedtranscriptionfactors,theircardseqandthecorresponding 247 singularsurfaces,ifanyAsannounced,intheselectedtranscriptionfactors,thereexistsa 248 correlationbetweenthelackof‘syntacticalfreedom’,orthepresenceofasingularsurface 249 inthecharactervariety,withanidentifieddisease. 250


3.3.AlgebraicmorphologyofmicroRNAs 251
MicroRNAs(miRNAs)playafundamentalroleintheexpressionandregulationof 252 genesbytargetingspecificmessengerRNAs(mRNAs)fordegradationortranslational 253 repression.ThemiRNAsareapproximately22ntlongsingle-strandedRNAmolecules. 254 ThegenesencodingmiRNAsaremuchlongerthantheprocessedmaturemiRNAmolecule. 255 ManymiRNAsareknowntoresideinintronsoftheirpre-mRNAhostgenesandsharetheir 256 regulatoryelements,primarytranscript,andhaveasimilarexpressionprofile.MicroRNAs 257 aretranscribedbyRNApolymeraseIIaslargeRNAprecursorscalledpri-miRNAs.Thepre- 258 miRNAsareaproximately70-nucleotidesinlengthandarefoldedintoimperfectstem-loop 259 structures,seeFigure 1 (Right)foranexample. 260
EachmiRNAissynthesizedasamiRNAduplexcomprisingtwostrands(-5pand-3p). 261 However,onlyoneofthetwostrandsbecomesactiveandisselectivelyincorporatedinto 262
VersionDecember13,2022submittedto Int.J.Mol.Sci. 9of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
theRNA-inducedsilencingcomplexinaprocessknownasmiRNAstrandselection[31,32]. 263 FordetailsaboutthemirRNAsequencesweusetheMirdatabase[33,34]. 264
PlantmiRNAsusuallyhavenear-perfectpairingwiththeirmRNAtargetssothat 265 generepressionproceedsthroughcleavageofthetargettranscripts.Incontrast,animal 266 miRNAsareabletorecognizetheirtargetmRNAsbyusingasfewas 6 to 8 nucleotides 267 (theseedregion)whichisnotenoughpairingforleadingtocleavageofthetargetmRNAs. 268 AgivenmiRNAmayhavehundredsofdifferentmRNAtargets,andagiventargetmight 269 beregulatedbymultiplemiRNAs. 270

DisregulationofmiRNAsmayleadtoadiseaselikecancer.AkeymicroRNAknown 271 asanoncommir(involvedinimmunityandcancer)ismir-155. 272
Specificallythe-3pstrandismir-155-3p.Figure 5 (top)illustratesthecomplementary 273 base-pairingbetweenmiR-155-3pandthehumanIRAK3(interleukin-1receptor-associated 274 kinase3)mRNA[10,Figure4]andtherelevantseedsequenceUCCUAC.Thecardseqfor 275 thissequenceisthetwo-letterfreefroup F2 andtheGroebnerbasisforthecorresponding 276 charactervarietycontainsthesurface f (A1) b (x, y, z)= x2 + y2 6z2 + 4xyz thathasasingle 277 simplesingularityasshownatthebottomofFigure 5.Ifoneretainsthefullseedsequence 278 isUCCUAC(A)thenthecardseqpassestothatofthefreegroup F2 tothegroup π2 andthe 279 singularsurfaceislost.Thisisacasewherethe‘bandwidth’oftheseediscriticalinthe 280 (dis)regulationofthemiRNA.TheseresultsaretranscribedinTable 3 281
Figure5. Up:Complementarybase-pairingbetweenmiR-155-3pandthehumanIrak3(interleukin-1 receptor-associatedkinase3)mRNA[10,Figure5].Therequisite‘seedsequence’base-pairingis denotedbythebolddashes.Down:thesurface f (A1 ) b (x, y, z)= x2 + y2 6z2 + 4xyz
Forthecaseof-5pstrandmir-155-3p,theseedsequenceUUAAUGCUAcontains 282 fourdistinctletters.ThiscaseissimilartogenericGroebnerbasesobtainedfromfour 283 letterseeds.Dependingonthechoicesofparameters a, b, c, d, e, f , g, h,theGroebnerbasis 284 containstheCayleycubic f (4) H (x, y, z),theFrickesurface κ3(x, y, z) (thatisrelatedtothe 285 linkL6a1[21,Figure2]),thesurface f (3A1) a (x, y, z) showninFigure 2 andothersurfaces.In 286 thisgenericcase,thesurfaceisfoundwith(atmost) 4 copieswhereeachcopyisattached 287 toadistinctpunctureofthe4-punctured4-sphere S4,2. 288 Intable 3,thisgenericcaseisdenoted 4 × generic (or 3 × generic formir-133-5p). 289 TheseresultsaretranscribedinTable 4. 290
AsmalllistofhumamiRNAsisinvestigatedinTables 3 and 4 correspondingto 3-letter 291 and 4-letterseeds.theprefix‘hsa’isforthehumanspecie.Likefortranscriptionfactors 292
VersionDecember13,2022submittedto Int.J.Mol.Sci. 10of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
Table3. Afewhuman(prefix‘hsa’)microRNAswhosegroupstructureisawayfromafreegroupor whoseGroebnerbasisofthe SL(2, C) charactervarietycontainsasingularsurface.Thesymbolmiris fortheidentificationintheMirdatabase[34],seedisfortheseedofthemiRNA,cardseqisforthe cardinalitysequenceofconjugacyclassesofsubgroupsofthegroupwhoseseedisthegenerator,sing istheidentificationofasingularsurfacewithintheGroebnerbasis,thelastcolumnnisforareference paperandthecorrespondingdisease[31].Thecardseqfor π1 and π′ 1 aregivenin[3,Table5].The cardseqfor π′ 2 is [1,3,7,34,139,931,5208,43867 ··· ].Forhsa-mir-124-1-3p,oneencounterstheFricke surface f (A1 ) 2,{} = xyz + x2 + y2 + z2 2y inthecharactervariety.
mirseedcardseqsingref&disease hsa-mir-193b-5pGGGGUU π1 nosing[31,34] GGGGUUU π′ 1 nosinglungcancer hsa-mir-155-3pUCCUAC F2 f (A1) b (x, y, z) [31,32,34] UCCUACA π2 nosingmultiplesclerosis hsa-mir-193a-5pGGGUCUU F2 f (A1) b (x, y, z) [31,34] breastcancer hsa-mir-223-5pGUGUAUU... hsa-mir-133-3pUUGGUC F2 f (3A1) b (x, y, z) [31,34] UUGGUCC π′ 2 nosingatrialfibrillation hsa-mir-124-3pAAGGCA F2 f (3A1) b , f (A1) 2,{} [34,35]
AAGGCAC.nosingAlzheimer’sdisease Table4. TheoppositestrandofthemicroRNAconsideredinTable 3.Theseedsequenceismade of 4 distinctbasesandthecorrespondingcardseqisthefreegroup F3 ofrank 3.TheGroebner basiscontains 4 copiesofthegenericcollectionofsurfaces κ4(x, y, z), f (3A1 ) (x, y, z), κ3(x, y, z),etc,as showninFigure 6,exceptforthe-5pstrandofmir-133wherethereareonly 3 copiesofthegeneric surfaces.
mirseedcardseqsingref&disease hsa-mir-193b-3pACUGGCC F3 4× generic[31,34] hsa-mir-155-5pUUAAUGCUA..[31,32,34] hsa-mir-193a-3pACUGGCC..[31,34] hsa-mir-223-3pGUCAGUU... hsa-mir-124-5pGUGUUCA... hsa-mir-133-5pGCUGGUA.3× generic.[34,35]
inTable 2,thelackof‘syntacticalfreedom’,ortheoccurrenceofasingularsurfaceinthe 293 charactervariety,issymptomaticofadisease. 294
4.Conclusion
295
Wefoundinthisworkthatasignatureofadiseasemaybegivenintermsofthe 296 groupstructureofaDNA/RNAsequenceandtherelatedcharactervarietyrepresenting 297 thegroup.TheDNAmotifofatranscriptionfactor,ortheseedofamicroRNA,defines 298 thegeneratorofagroup π.Assoonas π isawayfromafreegroup Fr (with r + 1 the 299 numberofdistinctbasesinthesequence)orthe SL(2, C) charactervariety G of π contains 300 singularsurfaceswithisolatedsingularities,apotentialdiseaseisonsight.Onewouldlike 301 tobemorepredictiveinidentifyingthepotentialdiseasewithpeculiargroupsorsingular 302 surfaces. 303
Firstofall,mostofthetime,thesurfacesencounteredinthecontextofTFsandmiRNAs 304 aredegree 3 delPezzo,incontrasttosurfacesobtainedfromotherDNAsequences,as 305 inEquation 4.Butthedegree 3 delPezzofamilyisveryrich.Forinstance,thesingular 306 surface f (A1) b = x2 + y2 6z2 + 4xyz (seeFigure 5)ispartofthecharactervarietyofTF 307
VersionDecember13,2022submittedto Int.J.Mol.Sci. 11of13
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022 doi:10.20944/preprints202212.0256.v1
Figure6. Up:Complementarybase-pairingbetweenmiR-155-5pandthehumanSpi1(spleenfocus formingvirusproviralintegrationoncogene)[10,Figure4].Therequisite‘seedsequence’base-pairing isdenotedbythebolddashes.Down(fromlefttoright):thesurfaces f (4) H = κ4(x, y, z), f (3A1 ) (x, y, z) and κ3(x, y, z),fourcopiesofthemarecontainedwithintheGroebnerbasisforthecharactervariety.


Pitx1(seeTable 2)andofmiRNAs155-3pand193a-5p(inTable 3).Then,thesingular 308 surface f (A1) 2,{} = x2 + y2 + z2 + xyz 2y ispartofthecharactervarietyofmirRNA133-3p. 309
Bothsurfaceshaveasimplesingularpointoftype A1 butdistinctsingularsubschemes(see 310 Section 2 forthenotation). 311

Anexceptiontothedegree 3 delPezzorulewasfoundininvestigatingthecharacter 312 varietyforthePrdm1transcriptionfactorinsubsection 3.1 313
Dothesefeaturesandotheronestobedescribedlatermayhelpforthediagnosticofa 314 potentialdisease?Thereisroomformuchworkinthefuturealongtheselines. 315 AuthorContributions: “Conceptualization,M.P.;methodology,M.PandM.A.;software,M.P.;vali- 316 dation,M.A.andK.I.;formalanalysis,M.P.;investigation,M.A.;resources,K.I.;datacuration,M.P.; 317 writing—originaldraftpreparation,M.P.;writing—reviewandediting,M.P.andM.A.;visualization, 318 M.A.;supervision,M.P.;projectadministration,K.I.;fundingacquisition,K.I.Allauthorshaveread 319 andagreedtothepublishedversionofthemanuscript”. 320
330 andgenomedecoding. Curr.IssuesMol.Biol. 2022, 44,1417–1433. 331
4. Dang,Y.;Gao,J.;Wang,J.;Heffernan,R.;Hanson,J.;Paliwal,K.;Zhou,Y.Sixty-fiveyearsofthelongmarchinproteinsecondary 332 structureprediction:Thefinalstrech? Brief.Bioinform. 2018, 19,482–494. 333
5. Bansal,A.;Kaushik,S.;Kukreti,S.Non-canonicalDNAstructures:Diversityanddiseaseassociation. Front.Genet. 2022, 13,959258. 334 https://doi.org/10.3389/fgene.2022.959258. 335
VersionDecember13,2022submittedto Int.J.Mol.Sci. 12of13
Funding:
InformedConsentStatement: “Notapplicable” 322 DataAvailabilityStatement: “Dataareavailablefromtheauthorsafterareasonabledemand” 323 ConflictsofInterest: “Theauthorsdeclarenoconflictofinterest” 324 MDPIMultidisciplinaryDigitalPublishingInstitute 325 References 326 1. Planat,M.;Aschheim,R.;Amaral,M.M.;Fang,F.;Irwin,K.Graphcoveringsforinvestigatingnonlocalstructuresinprotein, 327 musicandpoems. Science 2021, 3,39. 328 2. Irwin,K.Thecode-theoreticaxiom;thethirdontology. Rep.Adv.Phys.Sci. 2019
3
329 3. Planat,M.;Amaral,M.M.;FangF.;Chester,D.;Aschheim,R.;Irwin,K.GrouptheoryofsyntacticalfreedominDNAtranscription
“Thisresearchreceivednoexternalfunding” 321
,
,39.
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 December 2022
doi:10.20944/preprints202212.0256.v1
VersionDecember13,2022submittedto Int.J.Mol.Sci. 13of13
6. Planat,M.;Aschheim,R.;Amaral,M.M.;FangF.;Chester,D.;IrwinK.Charactervarietiesandalgebraicsurfacesforthetopology 336 ofquantumcomputing. Symmetry 2022,14,915. 337
7. Planat,M.;Amaral,M.M.;FangF.;Chester,D.;Aschheim,R.;Irwin,K.DNAsequenceandstructureundertheprismofgroup 338 theoryandalgebraicsurfaces. Int.J.Mol.Sci. 2022, 23,13290. 339
8. Gröbnerbasis.Availableonline:https://en.wikipedia.org/wiki/Gröbner_basis(accessedon1August2022). 340
9. Martinez,N.J.;Walhout,A.J.M.TheinterplaybetweentranscriptionfactorsandmicroRNAsingenome-scaleregulatorynetworks. 341 Bioessays 2009, 31,435–445. 342
10. miR-155.Availableonline:https://en.wikipedia.org/wiki/MiR-155(accessedon18November2022). 343
11. Kwak,J.H.;Nedela,R.Graphsandtheircoverings. Lect.NotesSer. 2007, 17,118. 344 12. TheOn-LineEncyclopediaofIntegerSequences.Availableonline:https://oeis.org/book.html(accessedon1November2022). 345 13. Hehl,F.W.;vonderHeyde,P.;Kerlick,G.D.;Nester,J.M.,Generalrelativitywithspinandtorsion:Foundationsandprospects. Rev. 346 Mod.Phys. 1976, 48 (3),393-416. 347
14. Rovelli,C.,Vidotto,F.:CovariantLoopQuantumGravity.CambridgeUniversityPress1edition,(2014). 348 15. Goldman,W.M.TracecoordinatesonFrickespacesofsomesimplehyperbolicsurfaces.In HandbookofTeichmüllertheory,Eur. 349 Math.Soc.,Zürich, 2009, 13,611-684. 350
16. Ashley,C.;BurelleJ.P.;Lawton,S.Rank1charactervarietiesoffinitelypresentedgroups, Geom.Dedicata 2018, 192,1–19. 351
17. Pythoncodetocomputecharactervarieties.Availableonline:http://math.gmu.edu/slawton3/Main.sagews(accessedon1May 352 2021). 353
18. Bosma,W.;Cannon,J.J.;Fieker,C.;Steel,A.(eds). HandbookofMagmafunctions,Edition2.23;UniversityofSydney:Sydney, 354 Australia,2017;5914pp(accessedon1January2019). 355
19. Cantat,S.BersandHénon,PainlevéandSchrödinger. 2009, DukeMath.J.149,411–460. 356
20. Farb,B.;Margalit,D. Aprimeronmappingclassgroups;PrincetonUniversityPress:Princeton,NewJersey,USA,2012. 357
21. Planat,M.;Chester,D.;Amaral,M.;IrwinK.Fricketopologicalqubits. Quant.Rep. 2022, 4,523-532. 358
22. Benedetto,R.L.;GoldmanW.M.Thetopologyoftherelativecharactervarietiesofaquadruply-puncturedsphere. Experiment. 359 Math. 1999, 8,85–103. 360
23. Iwasaki,K.Anarea-preservingactionofthemodulargrouponcubicsurfacesandthePainlevéVI. Comm.Math.Phys. 2003, 242, 361 185-219. 362
24. Inaba,M.;Iwasaki,K.;Saito,M.H.DynamicsofthesixthPainlevéequation. arXiv 2005,arXiv:math.AG/0501007 363
25. EnriquesKodairaclassification,availableonline:https://en.wikipedia.org/wiki/Enriques-Kodaira_classification 364
26. Doody,G.M.;Care,M.A.;Burgoyne,N.J.;Bradford,J.R.;Bota,M.;Bonifer,C.;Westhead,D.R..Anextendedsetof 365 PRDM1/BLIMP1targetgeneslinksbindingmotiftypetodynamicrepressionooze,R.M. Nucl.AcidsRes. 2010, 38 5336–5350. 366
27. Sandelin,A.;Alkema,W;Engström,P;Wasserman,WW;Lenhard,B,JASPAR:anopen-accessdatabaseforeukaryotictranscription 367 factorbindingprofiles. NucleicAcidsResearch 2004, 32,D91–D94;softwareavailableathttps://jaspar.genereg.net/(accessedon1 368 November2022). 369
28. Jauch,R.CrystaltructureandDNAindingofthehomeodomainofthestemcelltranscriptionfactorNanog. J.Mol.Biol. 2008, 376, 370 758–770. 371 29. Schuff,M;SiegelD.;Philipp,M.Bunsschu,K.;Heymann,N.;Donow,C.;Knöchel,W.CharacterizationofDaniorerioNanogand 372 FunctionalComparisontoXenopusVents. StemcellsandDevt. 2012, 21,1225–1238. 373 30. Schaeffer,L.N.;Huchet-Dymanus,M.;ChangeuxJ.P.ImplicationofamultisubunitEts-relatedtranscriptionfactorinsynaptic 374 expressionofthenicotinicacetylcholinereceptor. EMBOJ. 1998, 17,3078–3090. 375 31. Medley,C.M.;PanzadeG.;Zinovyeva,A.Y.MicroRNAstranselection,:unwindingtherules. WIREsRNA 2021, 12,e1627. 376 32. Dawson,O.;Piccinini,A.M.miR-155-3p:processingby-productorrisingstarinimmunityandcancer? OpenBiol. 2022, 12,220070. 377 33. Kozomara,A.;Birgaonu,M.;Griffiths-Jones,S.miRBase:frommicroRNAsequencestofunction. Nucl.AcidsRes. 2019, 47, 378 D155–D162. 379 34. miRBase:themicroRNAdatabase.Availableonline:https://www.mirbase.org/(accessedon1November2022). 380 35. Kou,X.;Chen,D.;Chen,N.TheregulationofmicroRNAsinAlzheimer’sdisease. Front.Neurol. 2020, 11,288. 381
| NOT PEER-REVIEWED | Posted:
View publication stats
Preprints (www.preprints.org)
14 December 2022 doi:10.20944/preprints202212.0256.v1

















