Mass Spectrometry-Based Approaches for Treating Human Diseases and Diagnostics
Editor Thiago Verano-Braga
Department of Physiology and Biophysics
Federal University of Minas Gerais
Belo Horizonte - MG, Brazil
ISSN 0065-2598
ISSN 2214-8019 (electronic)
Advances in Experimental Medicine and Biology
ISSN 2730-6216
ISSN 2730-6224 (electronic)
ISBN 978-3-031-50623-9 ISBN 978-3-031-50624-6 (eBook) https://doi.org/10.1007/978-3-031-50624-6
Proteomics, Metabolomics, Interactomics and Systems Biology
© The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG 2024
This work is subject to copyright. All rights are solely and exclusively licensed by the Publisher, whether the whole or part of the material is concerned, specifcally the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microflms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed.
The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specifc statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use.
The publisher, the authors, and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. Neither the publisher nor the authors or the editors give a warranty, expressed or implied, with respect to the material contained herein or for any errors or omissions that may have been made. The publisher remains neutral with regard to jurisdictional claims in published maps and institutional affliations.
This Springer imprint is published by the registered company Springer Nature Switzerland AG
The registered company address is: Gewerbestrasse 11, 6330 Cham, Switzerland
Paper in this product is recyclable.
Preface
Omics, a multidisciplinary feld encompassing genomics, transcriptomics, proteomics, metabolomics, lipidomics, and more, has witnessed remarkable growth in recent decades. This expansion owes much to the advancements in mass spectrometry (MS) instruments, innovative sample preparation techniques, and enhanced bioinformatics tools. Today, omic approaches are integral to the study of human diseases, driving transformative changes in the feld of Molecular Medicine.
This book delves into the dynamic landscape of molecular medicine, where mass spectrometry-based omic technologies, such as proteomics and metabolomics, are playing pivotal roles. These methodologies enable comprehensive molecular-level characterizations, paving the way for the discovery of novel biomarkers and therapeutic targets. The ultimate aspiration is personalized medicine, and omics offers valuable insights in this pursuit.
Within these pages, readers will encounter a wealth of information on the latest technological advancements in omics. The book navigates through the application of omics to gain deeper insights into human diseases, encompassing biomarker discovery, validation, and mechanistic understandings that can inform innovative treatments.
The initial chapters are dedicated to cutting-edge methods for isolating and characterizing extracellular vesicles (EVs) and introduce the innovative bioinformatic tool “Glycosort,” which enhances the post-processing of large-scale intact glycopeptide analyses. Subsequently, the book presents a series of review articles showcasing how omics approaches are illuminating various facets of human diseases, including those caused by viruses (SARS-CoV-2 and Zika Virus), psychiatric disorders, neurodegenerative diseases – including their impact in the intricate process of memory formation – and conditions affecting the cardiovascular, renal, and endocrine systems.
This book holds particular relevance for students, scientists, and healthcare professionals eager to explore the molecular intricacies of human diseases. It provides a valuable resource for those interested in identifying potential new diagnostic biomarkers and gaining a deeper understanding of disease progression.
Belo Horizonte, MG, Brazil Thiago Verano-Braga
1 Isolation of Extracellular Vesicles Using Titanium Dioxide Microspheres .
Veronica Feijoli Santiago, Livia Rosa-Fernandes, Janaina Macedo-da-Silva, Claudia B. Angeli, Simon Ngao Mule, Claudio R. F. Marinho, Ana Claudia Torrecilhas, Suely N. K. Marie, and Giuseppe Palmisano
2 Glycosort: A Computational Solution to Post-process Quantitative Large-Scale Intact Glycopeptide Analyses . . .
Lucas C. Lazari, Veronica Feijoli Santiago, Gilberto S. de Oliveira, Simon Ngao Mule, Claudia B. Angeli, Livia Rosa-Fernandes, and Giuseppe Palmisano
3 Applications of Mass Spectrometry in the Characterization, Screening, Diagnosis, and Prognosis of COVID-19 .
Camila Akemi Oliveira Yamada, Bruno de Paula Oliveira Santos, Rafael Pereira Lemos, Ana Carolina Silva Batista, Izabela Mamede C. A. da Conceição, Adriano de Paula Sabino, Luís Maurício Trambaioli da Rocha e Lima, and Mariana T. Q. de Magalhães
4 Proteomics and Metabolomics in Congenital Zika Syndrome: A Review of Molecular Insights and Biomarker Discovery
Patricia Sosa-Acosta, Fábio C. S. Nogueira, and Gilberto B. Domont
1
23
33
63
5 Mass Spectrometry Applications to Study Human Microbiome . . . . 87
Mohamed A. Raslan, Sara A. Raslan, Eslam M. Shehata, Amr S. Mahmoud, Marcus Vinicius Canário Viana, Flávia Aburjaile, Debmalya Barh, Nagwa A. Sabri, and Vasco Azevedo
6 Neuroproteomics: Unveiling the Molecular Insights of Psychiatric Disorders with a Focus on Anxiety Disorder and Depression . . . . . . . 103
Lucas Rodrigues-Ribeiro, Bruna Lopes Resende, Maria Luiza Pinto Dias, Megan Rodrigues Lopes, Larissa Luppi Monteiro de Barros, Muiara Aparecida Moraes, Thiago Verano-Braga, and Bruno Rezende Souza
7 Proteomic-Based Studies on Memory Formation in Normal and Neurodegenerative Disease-Affected Brains .
Ikaro Jesus da Silva Beraldo, Mateus Prates Rodrigues, Rafaela Schuttenberg Polanczyk, Thiago Verano-Braga, and Cleiton Lopes-Aguiar
129
8 Cardioproteomics: Insights on Cardiovascular Diseases 159
Brenda Raíssa-Oliveira, Ana Carolina Lara-Ribeiro, Júlia Rezende-Ribeiro, Ana Beatriz Queiroz Bahia, and Thiago Verano-Braga
9 Kidney Disease and Proteomics: A Recent Overview of a Useful Tool for Improving Early Diagnosis 173
Nicolly Emanuelle de Souza Barcelos, Maria Laura Limeres, Ana Flavia Peixoto-Dias, Maria Aparecida Ribeiro Vieira, and Diogo B. Peruchetti
10 Multi-omics Investigations in Endocrine Systems and Their Clinical Implications 187
Rodrigo Antonio Peliciari-Garcia, Carolina Fonseca de Barros, Ayla Secio-Silva, Diogo de Barros Peruchetti, Renata Marino Romano, and Paula Bargi-Souza
11 Omics to Unveil Diabetes Mellitus Pathogenesis and Biomarkers: Focus on Proteomics, Lipidomics, and Metabolomics 211
Nícia Pedreira Soares, Gabriela Castro Magalhaes, Pedro Henrique Mayrink, and Thiago Verano-Braga
12 Proteomics: Unraveling the Cross Talk Between Innate Immunity and Disease Pathophysiology, Diagnostics, and Treatment Options 221 Isabelle Souza Luz, Raquel Takaya, Daiane Gonzaga Ribeiro, Mariana S. Castro, and Wagner Fontes
13 Applications of Proteomics in Probiotics Having Anticancer and Chemopreventive Properties .
Mohamed A. Raslan, Sara A. Raslan, Eslam M. Shehata, Amr S. Mahmoud, Marcus Vinicius Canário Viana, Debmalya Barh, Nagwa A. Sabri, and Vasco Azevedo
243
14 Mass Spectrometry-Based Characterization of Protein Aggregates in Tissues and Biofluids
Janaina Macedo-da-Silva, Livia Rosa-Fernandes, Verônica Feijoli Santiago, Claudia Angeli Blanes, Suely Kazue Nagahashi Marie, and Giuseppe Palmisano
Index
Isolation of Extracellular Vesicles Using Titanium Dioxide Microspheres
Veronica Feijoli Santiago, Livia Rosa-Fernandes, Janaina Macedo-da-Silva, Claudia B. Angeli, Simon Ngao Mule, Claudio R. F. Marinho, Ana Claudia Torrecilhas, Suely N. K. Marie, and Giuseppe Palmisano
1.1 Introduction
1.1.1 Extracellular
Vesicle Biosynthesis and Biological Functions in Diseases
Extracellular vesicles (EVs) are characterized as nanoparticles enclosed by a lipid bilayer membrane secreted by the cells [1, 2]. The EVs can be classifed into three main types, according to the origin and size of two main types of EVs are distinguished based on their biogenesis, known as exosomes and ectosomes [3]. More specifcally; exosomes, microvesicles, and apoptotic bodies [3–5]. The exosomes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/978-3-031-50624-6_1.
V. F. Santiago · J. Macedo-da-Silva · C. B. Angeli · S. N. Mule · C. R. F. Marinho
Department of Parasitology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
L. Rosa-Fernandes
Department of Parasitology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
Centre for Motor Neuron Disease Research, Macquarie Medical School, Faculty of Medicine, Health and Human Sciences, Macquarie University, Sydney, Australia
A. C. Torrecilhas
Instituto de Ciências Ambientais, Químicas e Farmacêuticas; Departamento de Ciências Farmacêuticas; Laboratório de Imunologia Celular e Bioquímica de Fungos e Protozoários. Departamento de Ciências Farmacêuticas, Universidade Federal de São Paulo (UNIFESP), São Paulo, Brazil
© The Author(s), under exclusive license to Springer Nature Switzerland AG 2024 T. Verano-Braga (ed.), Mass Spectrometry-Based Approaches for Treating Human Diseases and Diagnostics, Advances in Experimental Medicine and Biology 1443, https://doi.org/10.1007/978-3-031-50624-6_1
V. F. Santiago et al.
are particles with 50–150 nm diameter, with endosomal biogenesis in the multivesicular bodies (MVBs) and amphisomes. The exosome’s biosynthesis occurs by inward budding endosomal vesicles driven by the endosomal sorting complex required for transport (ESCRT) machinery and fusion of these multivesicular bodies or amphisomes with the plasma membrane, resulting in the release of intraluminal vesicles (exosomes) [3, 6–8]. Microvesicles are medium-sized EVs (200–800 nm) that originated from cell membrane budding and are less abundant than small EVs [3, 9]. Microvesicles are released from many cells, such as neurons, platelets, red blood cells, astrocytes, and endothelial cells, in both healthy and pathological states [10, 11]. Apoptotic bodies are the largest EVs, comprising >1 μm released during the apoptotic process [12]. The heterogeneity of the EVs is related to the type, function, and state of the origin cell such as different mechanisms of cell death [3]. The EVs carry acid nucleic (DNA, mRNA, miRNA) proteins [13, 14], transcription factors, growth factors, signaling molecules, and lipids from one cell to another cell [15–19]. EVs are involved in several biological functions beyond the transport of molecules between cells, such as cell-cell communication and activation of target cells through EV surface ligands [20]. Moreover, EVs are involved in angiogenesis [21], infammation [22], coagulation [23], antigen presentation in immune response [24, 25], and fetal-maternal communication [26]. In diseases, EVs also participate in several biological processes. For example, in glioblastoma, cells secret mRNA, miRNA, and angiogenic proteins encapsulated in exosomes to target microvascular endothelial cells inducing angiogenesis [27]. EVs also participate in oncogenesis, cancer progression, and metastasis [28]. Isolated EVs have been described as potential prognostic and diagnostic tools in prostatic cancer [29, 30], ovarian cancer [31], lung adenocarcinoma [32], colon cancer [33], bladder cancer [11], breast cancer [34], and melanoma [35]. Moreover, EVs released from the brain were involved in Alzheimer’s disease mechanisms [36].
EVs are important in the host-pathogen interaction, being secreted both from the host and the pathogen. For example, parasites can deliver EVs that modulate the host’s immune response. Moreover, macrophages infected by Toxoplasma gondii release EVs, which are pro-infammatory [37]. Leishmania-delivered exosomes are involved in the modulation of macrophage/dendritic cells’ response to infection.
S. N. K. Marie
Laboratory of Molecular and Cellular Biology (LIM15), Department of Neurology, Fac-uldade de Medicina FMUSP, University of São Paulo, São Paulo, Brazil
G. Palmisano (*)
Department of Parasitology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
School of Natural Sciences, Faculty of Science and Engineering, Macquarie University, Sydney, Australia
e-mail: palmisano.gp@usp.br
Additionally, parasites can use EVs to evade host immunity. For example, Trypanosoma cruzi uses host microvesicles to evade host immunity [38–41]. Bacteria are also able to produce and release EVs, which are essential in the pathogenicity, cell-cell signaling between intra- and inter-species, and different organisms via transference of the EV content. In Gram-positive bacteria, the EVs are spherical bilayers with sizes between 20 and 400 nm [42, 43]. In Gram-negative bacteria, particles are described as outer membrane vesicles (OMVs) and display several roles, such as antibiotic resistance, nucleic acid transference, bioflm synthesis, and host-bacteria interaction [44, 45]. Specifcally, during infection, host cells can incorporate bacterial OMVs by endocytosis via different mechanisms: clathrindependent, caveolin-mediated, lipid raft, and membrane fusion [44]. Some inhibitors can be used to block OMV endocytosis, such as chlorpromazine (blocks clathrin-coated pit formation), dynasore (inhibits dynamin GTP activity in the caveolin-mediated endocytosis), wortmannin (inhibits the phosphatidylinositol kinases in the lipid raft endocytosis), and cytochalasin D (depolymerizes actin in the membrane fusion) [44]. Fungi EV’s content can activate the host immune system once recognized by pattern recognition receptors expressed by leukocytes [46]. Among the fungal EV cargo, described as virulence factors are lipids (phospholipids, sterols, glycolipids) [47, 48], proteins [47], polysaccharides [49], nucleic acids [50], and pigments such as melanin [51]. Among the proteins identifed in the EV cargo, urease and phosphatase were described in EV-released C. neoformans Urease is characterized in C. neoformans as a relevant virulence factor, involved in the host central nervous system invasion [52]. Fungi EVs are involved in adhesion in the brain microvasculature in C. neoformans [53] and stimulation of cytokine production by macrophage (IL-10, IL-12, TGF-β) and dendritic cells (IL-10, IL-12, TNF-α) in C. albicans [54]. EVs are also important in viral infections. Infected cells produce EVs that mediate the communication between infected and uninfected cells, supporting the virus survival and manipulating the host’s immune response [55].
The EVs’ molecular repertoire defnes their molecular and biological functions. Several reviews have investigated the analytical and computational methods to characterize EVs’ transcriptome, proteome, and metabolome in different pathophysiological conditions. Using mass spectrometry-based proteomics, in addition to the isolation and characterization of the proteins, it is possible to identify and characterize the EV proteins. Lischnig and colleagues [56] isolated large and small EVs using differential ultracentrifugation combined with density gradient centrifugation. In this study, more than 6000 proteins were identifed in EVs isolated from breast cancer cell lines (MDA-MB-231-luc-D3H1, MDA-MB-231luc-D3H2LN, and MDA-MB-231-luc-BMD2a), where 818 and 1567 proteins were enriched in small EVs and large EVs, respectively, isolated from all three cell lines. Most of the 1567 enriched proteins in large EVs were related to organelles such as mitochondrion, endoplasmic reticulum, and ribosomes, while the enriched proteins in small EVs were associated with multivesicular bodies, plasma membrane, and Golgi apparatus [56]. These results highlighted that the differences among the EV types are also at the protein content level. The EV proteome alterations are also observed during physical activities [57], where differences in
V. F. Santiago et al.
the EV proteins are released in the plasma over time points. The results showed regulation of 322 proteins among exercise and rest time points, mostly involved with multivesicular body formation, membrane traffcking, lipid rafts, vesicle adhesion, and chaperone [57]. EV proteome from B. anthracis [58], S. aureus [42], L. monocytogenes [59], P. acnes [60], and F. alocis [61] were analyzed using mass spectrometry-based proteomics to characterize the proteins involved in the EV release. The presence of penicillin-binding proteins (PBPs) and autolysins (transpeptidases related to cell wall homeostasis) as EV cargo suggests that the modifcation of the cell wall plays a role in the vesicle release. As in eukaryotes, bacterial EVs contain not only nucleic acids but also cytosolic and secreted proteins [44]. Metabolomics is described as other omics science potentially recommended to elucidate the infuence of cell metabolism in physical changes [62]. Metabolomics analyses were performed in urinary EVs from prostate cancer patients (PCa) and benign prostate hyperplasia (BPH). A total of 248 metabolites were identifed, including amino acids and lipids. In this comparison, 76 differentially regulated metabolites were identifed between PCa and BPH [63]. The regulated metabolites were mainly phosphatidylcholines (higher in BPH), sterols (higher in PCa), and fatty acid esters (higher in PCa). Moreover, vitamins were identifed as increased and glycerolipids decreased in PCa-released EVs. Interestingly, metabolite regulation changed according to PCa stages 2 and 3. In this context, three ceramides Cer(d18:1/16:0), Cer(d18:1/20:0), and Cer (d18:1/22:0), one glycerophospholipid (30:0), and stearoylcarnitine (18:0) showed different levels between stages 2 and 3. All these fndings support the urinaryreleased EVs as a non-invasive source for metabolite analysis in PCa [63]. Additionally, RNA, another EV cargo, has gained the interest of the study as a potential biomarker. Transcriptomic profling of plasma from multiple sclerosis patients showed a differential number and cargo of EVs. Using techniques such as differential ultracentrifugation and RNA-sequencing, EVs were isolated from 20 patients and 8 controls to identify differential expression of RNA, specifcally linear and circRNAs [64]. The main fnding was the identifcation of 6575 circRNAs in multiple sclerosis compared with healthy controls. Moreover, the study reported differences in the number of circRNAs and linear RNAs in the comparisons between multiple sclerosis and healthy controls or between different stages of the disease: relapsing-remitting vs secondary progressive multiple sclerosis (MS), highlighting the importance of further studies to analyze the function of RNAs in the progression of MS [64]. Transcriptomics analysis of EVs was also applied to infectious diseases. Meningher et al. [65] analyzed 26 schistosomiasispositive patients compared to 17 healthy controls and detected Schistosoma microRNAs in EVs isolated from host sera. All four schistosomal RNAs (bantam, miR-2c-3p, miR-3488, and miR-2a-5p) showed signifcant differences between the EVs isolated from infected and healthy individuals [65], suggesting the EVs as potential sources of diagnosis in schistosomiasis infection.
1.1.2 Omics Science Approaches for Extracellular Vesicle
Characterization
Innovative methods for EV proteome characterization have been developed and applied in healthy and diseased conditions [66]. The EVs’ proteome of human breast cancer subtype (triple-negative and HER2-positive) cell lines was characterized using ultracentrifugation with sequential centrifugation steps to remove cell debris and large EVs (10,000× g, 40 min), followed by two ultracentrifugation (UC) cycles at 120,000× g for 2 h each. This study identifed a total of 4992 proteins and 2299 differently regulated proteins (FDR 5%) using a mass spectrometry (MS) approach. Most of the proteins identifed in this study were reported in the EV databases ExoCarta and Vesiclepedia (>90%), and their biological processes and cellular component are related to cell-cell communication and EVs, respectively [67]. Mass spectrometry-based proteomics was applied to identify disease patterns and potential biomarkers for breast cancer by analyzing EV proteome. For this, Risha et al. (2020) [68] isolated EVs from a metastatic cancer cell line (MDA-MB-231) and a human breast epithelial cell line (MCF-10A) using three methods for isolation: ExoQuick, ultracentrifugation, and the combined ultrafltrationultracentrifugation. Among the three isolation methods, ExoQuick showed fve proteins, while UC and UF-UC presented 986 and 503 proteins, respectively. Using the UC method, the total proteome of MDA-MB-231 and MCF-10A identifed 1107 and 726 proteins, respectively. The gene ontology displayed extracellular exosomes as enriched cellular components. Moreover, 36 kinases were reported among the MDA-MB-231 exosomal proteins, and 3 exosomal membrane proteins (GLUT-1, Glypican-1, and ADAM-10) were hypothesized as a biomarker for breast cancer prognosis [68].
In addition to EV proteome analysis in several types of diseases, post-translational modifcations (PTMs) in EVs are also widely studied, focusing on the alteration of PTMs in cancers, pregnancy, and infection [69–71]. EVs can be isolated from several types of sources, such as cell culture and biofuids (bile, blood, urine, breast milk, semen, cerebrospinal fuid, saliva, and amniotic fuid) [72–76]. Due to this, there are many methods for EV isolation: UC, size exclusion chromatography (SEC), UF, density gradient, immunoaffnity, and microfuidics. Each method displays differences in specifcity, EV recovery, and purity. Each method requires a minimum sample input volume and shows different time and cost-associated procedures [77]. In some studies, the combination of more than one method is used to enrich the EVs. For example, ultrafltration (UF) combined with size exclusion chromatography (SEC) enhances the EVs’ isolation from cell culture media [78]. The EV isolation techniques and their technical challenges were reviewed by Ramirez et al. [79] and Stam et al. [80]. The gradient UC is another technique to separate aggregates from EVs using a sucrose gradient, where different particles foat in different densities (e.g., exosomes foatation 1.08–1.22 g/mL on a sucrose
V. F. Santiago et al.
gradient) [81, 82]. Moreover, the combination of differential ultracentrifugation followed by gradient UC (sucrose) was applied by Alexander et al. [83] to separate the contaminants. Among the isolation methods currently available, UC is the gold standard method [84]. In this technique, the isolation is based on the size of the particle. Larger vesicles are isolated by centrifugal force between 200 and 1000× g, while microvesicles and the exosomes form pellets by a centrifugal force of 10,000–20,000× g and 100,000× g, respectively [85–87]. The main disadvantage of UC is related to the low recovery of the EVs and time consumption, once the UC steps require 1–16 h of equipment use [88, 89]. Recently, Gao et al. [90] presented a novel isolation method for extracellular vesicles with high specifcity using metal oxide titanium dioxide (TiO2) microparticles. In EV isolation, the interaction occurs based on the reversible binding of TiO2 to the acidic phosphate groups present in the phospholipids in the EV bilayer membrane [90]. TiO2 microspheres are used to isolate phosphopeptides [91] and sialylated glycopeptides [92]. During the enrichment of these biomolecules, the pH of the loading solution is kept acidic to increase selectivity toward acidic residue-containing peptides. For the EV isolation, the pH of the loading solution is kept neutral [92]. This methodology allows EV isolation from both cell culture supernatants as also biofuids such as plasma, serum, and urine [93, 94]. Moreover, the TiO2 method showed a similar size profle of EVs compared to UC and smaller EVs compared to SEC fractions. Additionally, the time consumption is reduced in hours compared to UC and SEC methods [93]. The application of TiO2 was also effcient to analyze EVs isolated from the plasma of diabetic retinopathy (DR) patients to perform metabolomics analysis. The analysis compared diabetes mellitus (DM) and DR patients, stratifed into two groups: nonproliferative DR (NPDR, early stage) and proliferative DR (PDR, advanced stage). The results showed a total of 344 metabolites identifed using mass spectrometrybased proteomics. Several types of metabolites as amino acids, benzene, and aldehydes were identifed. Regulation of metabolites was observed in the comparison between NPDR and DM, where uracil, Ile-Phe, 4-acetamidobutyric acid, and MetHyp were upregulated, while lysophosphatidylcholine, sphingomyelins, and free fatty acid were downregulated in NPDR compared to DM. Taken together, these results highlight the relevant functions of EV metabolism during DR and its progression [93]. Considering the EV isolation from complex samples, such as plasma/ serum, the TiO2 enrichment method is described as a simple, rapid, low-cost, and low-volume sample required. Moreover, the technique shows a reduced plasma protein contamination (albumin, IgG) compared to the UC method, due to the high specifcity to isolate the EVs. During the EV isolation, the co-purifcation of the lipoproteins (ApoE, ApoB, ApoC-II, etc.) is also observed due to the similarities between these EVs and lipoproteins, such as the size and density, being a major challenge of the current techniques during the EV isolation from plasma samples [95–98]. Here, we present an adapted method for EV enrichment using TiO2 microspheres with low plasma volume and a step-by-step protocol, focusing on MALDI and LC-MS/MS proteomics approaches. An overview of the EV enrichment by the TiO2 method is represented in Fig. 1.1.
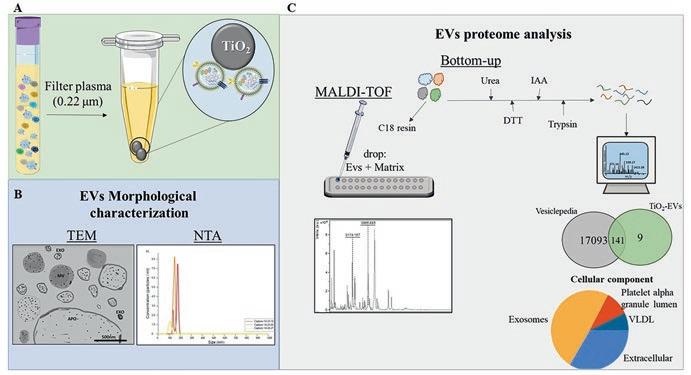
Fig. 1.1 Schematic representation of EV isolation method using TiO2 microspheres. (a) 100 μl of plasma was fltered using a 0.22-μm syringe flter with GHP Membrane (PALL Life Sciences, USA) directly into the 5 mg TiO2 bead microtube, incubating for 5 minutes at 4 °C, max speed at thermomixer. Three washing steps with fltered PBS are required to remove the non-EV molecules. The elution of EVs is performed with 10% ammonium hydroxide, pH 11.3. (b) The morphological characterization of EVs can be performed using transmission electron microscopy (TEM) and nanotracking analysis (NTA). Moreover, the EV proteome analysis is performed by mass spectrometry-based proteomics, MALDI-TOF, and LC-MS/MS
1.2 Material and Methods
1.2.1
Plasma Samples
Peripheral blood was collected at delivery into vacuum tubes containing heparin. Plasma was isolated by centrifugation at 1200× g for 10 min at 4 °C and maintained at 80 °C until use. Human plasma samples from healthy controls were supported by ethical clearance provided by the Ethic Committee in Research Involving Human Beings from the Biomedical Sciences Institute of the São Paulo University under the Certifcate of Presentation to the Ethic Appreciation (CAAE) numbers 03930812.8.0000.5467 and 32707720.0.0000.5467, according to the Resolution No. 466/12 from the Brazilian National Health Committee.
1.2.2 Enrichment of Plasma EVs Using TiO2 Microspheres
Add 5 mg of TiO2 beads (Titansphere TiO, 10 μm, GL Sciences) into the low binding microtube. Thaw plasma samples on ice. Then, mix them by vortexing followed by spinning down before fltering 100 μl of plasma using a 0.2-μm syringe flter
V. F. Santiago et al.
with GHP Membrane (PALL Life Sciences, USA) directly into the TiO2 bead microtube. Plasma was mixed with TiO2 beads by vortexing roughly for 40 s and incubated in a thermomixer system at 2000 RPM for 5 min at 4 °C. The beads were spun down for 1 min using a bench centrifuge, and remove the supernatant, once the EVs are adsorbed by TiO2 beads. TiO2 beads were washed three times briefy with 200 μl of fltered phosphate-buffered saline (PBS) solution, mixed by vortexing, and incubated in a thermomixer system at 2000 RPM for 5 min at 4 °C. The beads were spun down to remove the maximum volume of PBS solution before the EV elution step to avoid alterations in the elution buffer’s fnal concentration. The EV elution was performed by adding 50 μl of fresh elution buffer, which consists of 10% ammonium hydroxide (pH 11.3), and incubation in a thermomixer system at 2000 RPM for 15 min at 4 °C. Then, centrifuge for 10,000× g for 10 min at 4 °C to pellet the TiO2 beads, and collect the EVs in the supernatant. The following steps can vary according to the next analysis (MALDI-TOF MS, LC-MS/MS, etc.).
1.2.3 EV Morphological Characterization by Transmission
Electron Microscopy and Nanoparticle Tracking Analysis (NTA)
Below are reported detailed procedures for the morphological and molecular characterization of the isolated EVs. The procedures can be adapted based on the equipment and techniques used.
The 10% ammonium water needs to be exchanged for a proper solution preparing the Amicon® Ultra Centrifugation Filters 30 kDa cut-off (Merck Millipore, Billerica, MA, USA) by adding 200 μl of fltered PBS, and centrifuge 10,000× g for 1 min at 4 °C. Then, the eluted EV sample was added to the Amicon flter, and ammonium hydroxide was diluted 10x by adding PBS to complete the volume. The ammonium hydroxide was removed by centrifugation at 10,000× g for 5 min at 4 °C. The recovery of EVs was performed by inverting the Amicon flter into a new tube, and centrifuge 10,000× g for 5 min at 4 °C. The recovered EVs were placed into a new LoBind tube, and store the samples at 4 °C until further analysis.
1.2.3.1 For NTA Analysis
The EV samples were ten times diluted by fltered sterile PBS and injected in the sample chamber with sterile insulin syringes (BD, New Jersey, USA) until the liquid reached the tip of the nozzle. All measurements are performed at room temperature, with samples being kept on ice between analyses. The software used for capturing and analyzing the data is NTA 3.4. For the NTA analysis, a 500-μL volume of the isolated EV sample was manually injected into the laser chamber of the NanoSight NS300 instrument. The injection was carefully carried out to prevent any
introduction of air bubbles or sample loss. Readings were taken in triplicate for each sample, with each reading lasting 30 s at a frame rate of 25 frames per second. This allowed for the tracking and measurement of EVs in real time, capturing their Brownian motion. NTA was set up to three measurements for each sample for 30 s per recording, with manual shutter and gain adjustments, and kept for all samples. A value between 2.0 and 3.0 for screen gain and 5.0 and 6.0 camera level was selected for the analysis and kept for the following samples. The supplied data is used to measure the number of particles, the average and median size of vesicles obtained, and mode, D10, D50, and D90, comparing the profles between the groups.
1.2.3.2 For TEM Analysis
The EVs were fxed with 0.2% paraformaldehyde (w/v) for at least 18 h. Then, 10 μL of samples was incubated for 7 min in a 300-mesh grid with carbon-coated Formvar flm ((Electron Microscopy Sciences, Hatfeld, USA). The grid was incubated on 10 μL of 1.75% uranyl acetate (w/v) for 7 min. The samples were dried at room temperature for 2 h and imaged with TEM at 100 kV (CM-10, Philips, Eindhoven, the Netherlands).
1.2.4 EV Plasma Proteome Identifcation Using MALDI-TOF MS and LC-MS/MS
1.2.4.1 For the MALDI-TOF MS Approach
• The recovered EVs were lysed by adding 50 μL of 50% acetonitrile in 1% trifuoroacetic acid’s fnal concentration. Reserve 10% of the total volume for protein quantifcation (Qubit/Nanodrop), and dry down the lysed EV vacuum centrifuge.
• Resuspend the dried sample in 1% trifuoroacetic acid, and pass it through polymeric resin disks C18.
• Prepare the matrix solution (α-cyano-4-hydroxycinnamic acid [HCCA]) by dissolving in acetonitrile/water 50:50 vol/vol containing 2.5% trifuoroacetic acid at 10 mg/ml, mixed with the sample, directly spotted in duplicate onto a stainless steel MALDI target plate (Bruker Daltonics), and analyzed in a MALDI-TOF Autofex speed smart beam mass spectrometer (Bruker Daltonics) using FlexControl software (version 3.3.108.0; Bruker Daltonics).
• MALDI-TOF Autofex Setup: Spectra recording in the positive linear mode (laser frequency, 500 Hz; extraction delay time, 390 ns; ion source 1 voltage, 19.5 kV; ion source 2 voltage, 18.4 kV; lens voltage, 8.5 kV; mass range, 2400–20,000 Da). Spectra acquisition: automatic run mode to avoid subjective interference with the data acquisition. For each sample, 5000 shots, in 500-shot steps, were summed. All spectra calibration: using Protein Calibration Standard 1 Isolation of Extracellular Vesicles Using Titanium Dioxide Microspheres
V. F. Santiago et al.
I (Insulin [M + H]+ = 5734.52, Cytochrome C [M+ 2H]2+ = 6181.05, Myoglobin [M+ 2H]2+ = 8476.66, Ubiquitin I [M + H]+ = 8565.76, Cytochrome C [M + H]+ = 12,360.97, Myoglobin [M + H]+ = 16,952.31) (Bruker Daltonics). EV spectra are log-transformed using the square root method, followed by spectra smoothing using the Savitzky-Golay method and a half window size of 10. A base correction is required using the TopHat method. The intensity should be normalized using the total ion current method and peak identifcation with a halfwindow size of 10 and signal-to-noise ratios of 5 and 10. Raw data were imported to MALDIquantForeign [99] package and processed using the MALDIquant package [99] for TiO2-enriched analyses.
1.2.4.2 For LC-MS/MS Analysis
• Before adding the EVs into the 30 kDa Amicon flter, wash the membrane with 200 μl of fltered PBS, and centrifuge 10,000× g for 1 min at 4 °C.
• Add the eluted EV sample to a 30 kDa Amicon flter, and complete the volume with PBS to dilute ten times the ammonium hydroxide. Centrifuge 10,000× g for 5 min at 4 °C.
• Recover the concentrated sample by inverting the Amicon flter into a new tube, and centrifuge 10,000× g for 5 min at 4 °C.
• Collect the recovered EVs into a new LoBind tube, and add 4 M urea with a cocktail protease inhibitor for sonication for 1 h in the ice bath to lysis the EV membrane.
• Protein quantifcation was performed before adding 10 mM dithiothreitol (DTT) fnal concentration in 50 mM ammonium bicarbonate to the samples and incubating at 30 °C for 45 min.
• Add 40 mM iodoacetamide in 50 mM ammonium bicarbonate (fnal concentration), incubate for 30 min at room temperature, and be protected from the light.
• To quench the iodoacetamide alkylation, add 5 mM DTT fnal concentration, and incubate for 15 min at 30 °C.
• Before adding the trypsin (1:50, w/w), dilute ten times the urea with 50 mM ammonium bicarbonate, and confrm that the pH is between 7 and 8.
• Add trypsin and incubate for 16 h, at 30 °C, and 400 RPM on a thermoshaker. After this time, add 1% trifuoroacetic acid fnal concentration to stop the trypsin reaction. Check the pH (should be below 3).
• Centrifuge the samples for 10 min at 10.000× g, recover the supernatant, and proceed with the desalting protocol.
• For desalting protocol, insert two disks C18 microcolumn placed to a P200 pipette tip.
• Add 100 μl of activation solution (acetonitrile (PA) in the C18 resin, followed by 100 μl of 0.1% trifuoroacetic acid (TFA).
• Load peptides in the C18 column, and wash them three times with 0.1% TFA.
• Elute the peptides with 50% acetonitrile/0.1% trifuoroacetic acid followed by 70% acetonitrile/0.1% trifuoroacetic acid.
• Dry the samples in a vacuum concentrator, and store them in a 20 °C freezer until LC-MS/MS analysis.
1.2.5 Bioinformatics Analysis of Mass Spectrometry-Based Proteomics (LC-MS/MS)
• Resuspend the samples in 0.1% formic acid, and load on ReproSil-Pur C18 AQ (Dr. Maisch, Ammerbuch-Entringen, Germany) in-house packed trap column (2 cm × 100 μm inner diameter 5 μm) by reverse-phase chromatography, operated on an EASY-nLC II system (Thermo Scientifc) and fow 300 nL/minute.
Mobile phase water/0.1% FA as solvent A and solvent B (95% ACN/0.1% FA).
Gradient 5–28% of solvent B for 25 min, 28–40% of B for 3 min, and 40–95% of B for 2 min. The EASY-nLC II is coupled into an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientifc) and operates in positive mode ion. The mass spectrometer acquired a full MS scan at 120,000 full-width half maximum (FWHM) resolution with a 350–1550 Da mass range. The top 20 most intense peptide ions should be selected from MS for high energy collision-induced dissociation (HCD) fragmentation (collision energy: 30.0 V).
• The MS output fles are imported to protein identifcation into MaxQuant software [100]. In MaxQuant, for protein identifcation, the database search engine Andromeda was used against UniProt Homo sapiens (20,361 entries released). The following parameters can be used: carbamidomethylation of cysteine (57.021464 Da) as a fxed modifcation, oxidation of methionine (15.994915 Da), and N-terminal acetylation protein (42.010565 Da) as variable modifcations. Full trypsin with a maximum of two missed cleavages as enzyme specifcity. The minimum peptide length can be set to seven amino acids. For label-free quantifcation, use the “match between runs” feature in MaxQuant, which can identify the transfer between the samples based on the retention time and accurate mass, with a 0.7-min match time window and 20-min alignment time window.
1.3 Results of the EV Morphological Characterization and Proteome Identifcation
The results shown below were obtained from a plasma sample. The size distribution of the isolated EVs was obtained by NTA analysis. This technique is based on the Brownian motion using Stokes-Einstein equation to determine the nanoparticle size and concentration [101]. TiO2-enriched EVs showed a size means and mode size of
V. F. Santiago et al.
Fig. 1.2 TiO2-enriched EV characterization based on the size and concentrations measured by NTA. 500 μL from the isolated EV sample was manually injected into the laser chamber. Triplicate readings were taken for each sample, with each reading lasting 30 s at a frame rate of 25 frames per second. This allowed for real-time tracking and measurement of EVs by capturing their Brownian motion
Fig. 1.3 MALDI-TOF MS of the TiO2-enriched EVs
140.2 nm (±17.5 nm) and 144.4 nm (±20.2 nm), respectively. Moreover, the EV concentration obtained was 1.28e+07 particles/ml (Fig. 1.2).
EV proteome identifcation was performed using MALDI-TOF MS and LC-MS/ MS proteomics approaches. Figure 1.3 shows the TiO2-enriched EVs’ MALDI spectra. EV content showed more peaks established in the region 2000–6000 m/z with the most intense peaks localized around 2000, 3000, and 4000 m/z regions.
Complementary to MALDI-TOF MS proteomics, bottom-up proteomics using LC-MS/MS was performed to identify the proteins in the TiO2-enriched EVs. A total of 172 proteins were identifed in TiO2-enriched EVs (Supplementary Table 1.1). Based on the gene name, a comparison between the identifed proteins and the Vesiclepedia database was performed, showing an overlap of 90.7% of the EV proteins reported in the Vesiclepedia (Fig. 1.4a, Table 1.1). Among the reported EV proteins in Vesiclepedia are Protein S100-A8, Protein S100-A9, Transgelin-2,
1 Isolation of Extracellular Vesicles Using Titanium Dioxide Microspheres
Fig. 1.4 (a) Venn diagram of identifed proteins in TiO2-enriched EVs reported in Vesiclepedia. (b) Gene ontology of the EV proteins reported in Vesiclepedia
Caspase-7, Talin-1, disintegrin and metalloproteinase domain-containing protein 12 (ADAM-12), destrin, tetranectin, Ras suppressor protein 1, Coronin-1C, transmembrane protease serine 6, and Cystatin-C.
Regarding the gene ontology, the TiO2-EV proteins reported in Vesiclepedia showed Reactome pathways related to platelet activation, regulation of insulin-like growth factor and uptake by IGFBPs, regulation of complement cascade, and neutrophil degranulation. The biological processes associated with the EV proteins were proteolysis, blood coagulation, complement activation, negative regulation of endopeptidase activity, and cell adhesion. Additionally, molecular functions of EV protein were serine-type endopeptidase inhibitor activity, antigen binding, extracellular matrix structural constituent, phospholipid binding, and integrin binding. Regarding the cellular components, the external side of the plasma membrane, early endosome, extracellular vesicle, and endocytic vesicle lumen were the most enriched of the EV proteins (Fig. 1.4b).
1.4 Conclusion
The application of the TiO2 beads for EV enrichment of complex samples, such as plasma, is the most rapid and effective approach using a low volume of samples compared to the UC methods. Short time consumption enables the preparation of a high number of samples, allowing the application in the medical diagnosis. In this method, TiO2-enriched EVs showed a high concentration (1.28e+07 particles/ml) in NTA analysis, where the particle size was between 140 and 144 nm. Moreover, the
Table 1.1 EV proteins reported in Vesiclepedia
Ribonuclease pancreatic
Inter-alpha-trypsin inhibitor heavy chain H2
Hyaluronan-binding protein 2
Ceruloplasmin
Complement C1q subcomponent subunit C
Ras suppressor protein 1
Inter-alpha-trypsin inhibitor heavy chain H4
Hemoglobin subunit alpha
Carboxypeptidase N catalytic chain
Complement C1r subcomponent
Protein S100-A8
Protein S100-A9
Serum amyloid A-4 protein
Alpha-1-antitrypsin
Alpha-1-antichymotrypsin
Intelectin-1
Haptoglobin
Complement C1s subcomponent C-reactive protein
Plasma kallikrein
Haptoglobin-related protein
Cysteine and glycine-rich protein 1
Complement C3
Kininogen-1
Hemopexin
Cystatin-C
C4b-binding protein alpha chain
Apolipoprotein(a)
Histidine-rich glycoprotein
Neutrophil defensin 3
C4b-binding protein beta chain
Leucine-rich repeat and fbronectin type III domain-containing protein 1
Insulin-like growth factor II
Antithrombin-III
Heparin cofactor 2
V. F. Santiago et al.
Pigment epithelium-derived factor
Lysozyme C
Insulin-like growth factor-binding protein 1
Microtubule-associated protein RP/EB family member 2
Insulin-like growth factor-binding protein 2
Alpha-1-acid glycoprotein 1
Insulin-like growth factor-binding protein 3
Destrin
Complement component C6
Coagulation factor X
Complement component C8 alpha chain
Coagulation factor XI
Complement component C8 beta chain
Coagulation factor XII
Complement component C8 gamma chain
Albumin
Alpha-1B-glycoprotein
Alpha-2-macroglobulin
Actin, cytoplasmic 2
Disintegrin and metalloproteinase domain-containing protein 12
Afamin
Angiotensinogen
Alpha-2-HS-glycoprotein
Protein AMBP
Serum amyloid P-component
1 Isolation of Extracellular Vesicles Using Titanium Dioxide Microspheres
Ribonuclease pancreatic
Inter-alpha-trypsin inhibitor heavy chain H2
Hyaluronan-binding protein 2
Ceruloplasmin
Complement C1q subcomponent subunit C
Alpha-2-antiplasmin
Plasma protease C1 inhibitor
Sex hormone-binding globulin
Antileukoproteinase
Secreted phosphoprotein 24
TATA box-binding proteinassociated factor RNA polymerase I subunit A
Transgelin-2
Serotransferrin (continued)
Albumin
Alpha-1-acid glycoprotein 2
Insulin-like growth factor-binding protein 4
Procollagen C-endopeptidase enhancer 1
Insulin-like growth factor-binding protein 5
Protein disulfde-isomerase A3
Immunoglobulin heavy constant alpha 1
Plasminogen
Immunoglobulin heavy constant alpha 2
Serum paraoxonase/ arylesterase 1
Platelet basic protein
Immunoglobulin heavy constant gamma 1
Immunoglobulin heavy constant gamma 2
Peptidyl-prolyl cis-trans isomerase A
Peptidyl-prolyl cis-trans isomerase B
Immunoglobulin heavy constant gamma 3
Immunoglobulin heavy constant gamma 4
Prothrombin
Complement component C9
Apolipoprotein A-I
Coagulation factor VII
Adenylyl cyclaseassociated protein 1
Apolipoprotein A-II
Coagulation factor IX
Caspase-7
Apolipoprotein A-IV
Fibulin-1
CD5 antigen-like
Apolipoprotein B-100
Ficolin-1
Complement factor B
Apolipoprotein C-I
Fermitin family homolog 3
Complement factor D
Apolipoprotein C-II
Fetuin-B
Complement factor H
Fibrinogen alpha chain
Complement factor H-related protein 1
Apolipoprotein C-III
Apolipoprotein D
Table 1.1 (continued)
Ribonuclease pancreatic
Inter-alpha-trypsin inhibitor heavy chain H2
Hyaluronan-binding protein 2
Ceruloplasmin
Complement C1q subcomponent subunit C
Transforming growth factor-beta-induced protein ig-h3
Thrombospondin-1
Thrombospondin-4
Proteoglycan 4
Immunoglobulin heavy constant mu
Fibrinogen beta chain
Complement factor H-related protein 2
Vitamin K-dependent protein C
Immunoglobulin heavy variable 3–7
Fibrinogen gamma chain
Complement factor H-related protein 5
Vitamin K-dependent protein S
Immunoglobulin kappa constant
Intelectin-1
Putative pregnancy-specifc beta-1-glycoprotein 7
Immunoglobulin kappa variable 1–5
Transmembrane protease serine 6
Thymosin beta-4
Prostaglandin-H2 D-isomerase
Immunoglobulin kappa variable 3–20
Pregnancy zone protein
Immunoglobulin lambda constant 1
Transthyretin
Retinoic acid receptor responder protein 2
Immunoglobulin lambda constant 3
Fibroleukin
Four and a half LIM domains protein 1
Filamin-A
Fibronectin
Vitamin D-binding protein
V. F. Santiago et al.
Albumin
Apolipoprotein E
Beta-2-glycoprotein 1
Coflin-1
Properdin
Apolipoprotein L1
Zinc-alpha-2-glycoprotein
Tetranectin
Beta-2-microglobulin
Clusterin
Bridging integrator 2
Collagen alpha1(XVIII) chain
Complement C1q subcomponent subunit A
high specifcity of the interaction between EVs and TiO2 beads allowed the identifcation of EV proteins by LC-MS/MS, which their ions are usually suppressed by the most abundant plasma proteins (IgGs, albumin, etc.) and lipoproteins, chylomicrons, VLDL, LDL, and HDL. In conclusion, the combination of the TiO2 enrichment method applied for EV isolation and high-throughput mass spectrometry techniques allowed the EV characterization and proteome profling from EVs of a complex and viscous biofuid in an easy and fast way.
Competing Interests The authors declare no competing interests.
Supplementary Material The supplementary material can be downloaded from 10.6084/m9. fgshare.24194895.
Funding We are grateful for the fnancial support provided by the São Paulo Research Foundation (FAPESP, grant process nos. 2018/18257-1 (GP), 2018/15549-1 (GP), 2020/04923-0 (GP), 2021/00140-3 (JMDS), 2018/20468-0 and 2020/06747-4 (CRFM), 2017/04032-5 and 2021/14751-4 (SNM)), by the Conselho Nacional de Desenvolvimento Científco e Tecnológico (grant no. 870219/1997-9 (VFS) and “Bolsa de Produtividade” (CRF, SKNM, and GP)), by Fundação Faculdade de Medicina (FFM-SKNM), and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES bolsa PNPD to LRF), by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grant nos. 2019/15909-0 and 2020/07870-4 (ACT), and by Conselho Nacional de Desenvolvimento Científco e Tecnológico (CNPq, grant no. 408186/2018-6 (ACT)).
References
1. Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4):373–383
2. Théry C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2(8):569–579
3. Buzas EI (2023) The roles of extracellular vesicles in the immune system. Nat Rev Immunol 23(4):236–250
4. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1):1535750
5. Schiffelers R, Kooijmans S, Vader, van Dommelen, Van Solinge. (2012) Exosome mimetics: a novel class of drug delivery systems. IJN 7:1525
6. Qin J, Xu Q (2014) Functions and application of exosomes. Acta Pol Pharm 71(4):537–543
7. Colombo M, Raposo G, Théry C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30(1):255–289
8. Hessvik NP, Llorente A (2018) Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75(2):193–208
9. Silva M, A. Melo S. (2015) Non-coding RNAs in exosomes: new players in cancer biology. Curr Genomics 16(5):295–303
10. Revenfeld ALS, Bæk R, Nielsen MH, Stensballe A, Varming K, Jørgensen M (2014) Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 36(6):830–846
V. F. Santiago et al.
11. Smalley DM, Sheman NE, Nelson K, Theodorescu D (2008) Isolation and identifcation of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res 7(5):2088–2096
12. Atkin-Smith GK, Tixeira R, Paone S, Mathivanan S, Collins C, Liem M et al (2015) A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun 6(1):7439
13. Abels ER, Breakefeld XO (2016) Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 36(3):301–312
14. Witwer KW (2015) Circulating MicroRNA biomarker studies: pitfalls and potential solutions. Clin Chem 61(1):56–63
15. Yáñez-Mó M, Siljander PRM, Andreu Z, Zavec AB, Borràs FE, Buzas EI et al (2015) Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4:27066
16. Tkach M, Théry C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164(6):1226–1232
17. Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D et al (2014) Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to infammation. PLoS Biol 12(6):e1001874
18. Dang VD, Jella KK, Ragheb RRT, Denslow ND, Alli AA (2017) Lipidomic and proteomic analysis of exosomes from mouse cortical collecting duct cells. FASEB J 31(12):5399–5408
19. Doyle L, Wang M (2019) Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cell 8(7):727
20. Théry C, Duban L, Segura E, Véron P, Lantz O, Amigorena S (2002) Indirect activation of naïve CD4+ T cells by dendritic cell–derived exosomes. Nat Immunol 3(12):1156–1162
21. Todorova D, Simoncini S, Lacroix R, Sabatier F, Dignat-George F (2017) Extracellular vesicles in angiogenesis. Circ Res 120(10):1658–1673
22. Babuta M, Szabo G (2021) Extracellular vesicles in infammation: focus on the microRNA cargo of EVs in modulation of liver diseases. J Leukoc Biol 111(1):75–92
23. Berckmans RJ, Lacroix R, Hau CM, Sturk A, Nieuwland R (2019) Extracellular vesicles and coagulation in blood from healthy humans revisited. J Extracell Vesicles 8(1):1688936
24. Aharon A, Brenner B (2009) Microparticles, thrombosis and cancer. Best Pract Res Clin Haematol 22(1):61–69
25. Lindenbergh MFS, Stoorvogel W (2018) Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu Rev Immunol 36(1):435–459
26. Buca D, Bologna G, D’Amico A, Cugini S, Musca F, Febbo M et al (2020) Extracellular vesicles in feto–maternal crosstalk and pregnancy disorders. IJMS 21(6):2120
27. Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT et al (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476
28. Chang WH, Cerione RA, Antonyak MA (2021) Extracellular vesicles and their roles in cancer progression. Methods Mol Biol 2174:143–170
29. Øverbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V et al (2015) Identifcation of prostate cancer biomarkers in urinary exosomes. Oncotarget 6(30):30357–30376
30. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefeld XO et al (2009) Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 100(10):1603–1607
31. Lucidi A, Buca D, Ronsini C, Tinari S, Bologna G, Buca D et al (2020) Role of extracellular vesicles in epithelial ovarian cancer: a systematic review. IJMS 21(22):8762
32. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN et al (2013) microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 8(9):1156–1162
33. Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K et al (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9(4):e92921
34. Ekström K, Crescitelli R, Pétursson HI, Johansson J, Lässer C, Olofsson BR (2022) Characterization of surface markers on extracellular vesicles isolated from lymphatic exudate from patients with breast cancer. BMC Cancer 22(1):50
35. Suárez H, Andreu Z, Mazzeo C, Toribio V, Pérez-Rivera AE, López-Martín S et al (2021) CD9 inhibition reveals a functional connection of extracellular vesicle secretion with mitophagy in melanoma cells. J Extracell Ves [Internet] [cited 2023 July 16] 10(7):e12082. Available from: https://onlinelibrary.wiley.com/doi/10.1002/jev2.12082
36. Huang Y, Driedonks TAP, Cheng L, Rajapaksha H, Routenberg DA, Nagaraj R et al (2022) Brain tissue-derived extracellular vesicles in Alzheimer’s disease display altered key protein Levels including cell type-specifc markers. J Alzheimers Dis 90(3):1057–1072
37. Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS (2007) Exosomes released from macrophages infected with intracellular pathogens stimulate a proinfammatory response in vitro and in vivo. Blood 110(9):3234–3244
38. Cestari I, Ansa-Addo E, Deolindo P, Inal JM, Ramirez MI (2012) Trypanosoma cruzi immune evasion mediated by host cell-derived microvesicles. J Immunol 188(4):1942–1952
39. Torrecilhas AC, Soares RP, Schenkman S, Fernández-Prada C, Olivier M (2020) Extracellular vesicles in Trypanosomatids: host cell communication. Front Cell Infect Microbiol [Internet] [cited 2022 Oct 15] 10. Available from: https://www.frontiersin.org/articles/10.3389/ fcimb.2020.602502
40. Torrecilhas AC, Schumacher RI, Alves MJM, Colli W (2012) Vesicles as carriers of virulence factors in parasitic protozoan diseases. Microbes Infect 14(15):1465–1474
41. Trocolitorrecilhas A, Tonelli R, Pavanelli W, Dasilva J, Schumacher R, Desouza W et al (2009) Trypanosoma cruzi: parasite shed vesicles increase heart parasitism and generate an intense infammatory response. Microbes Infect 11(1):29–39
42. Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S et al (2009) Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureusderived membrane vesicles. Proteomics 9(24):5425–5436
43. Prados-Rosales R, Brown L, Casadevall A, Montalvo-Quirós S, Luque-Garcia JL (2014) Isolation and identifcation of membrane vesicle-associated proteins in gram-positive bacteria and mycobacteria. MethodsX 1:124–129
44. O’Donoghue EJ, Krachler AM (2016) Mechanisms of outer membrane vesicle entry into host cells: MicroReview – OMV entry into host cells. Cell Microbiol 18(11):1508–1517
45. Briaud P, Carroll RK (2020) Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect Immun 88(12):e00433-20
46. Patin EC, Thompson A, Orr SJ (2019) Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol 89:24–33
47. Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM et al (2008) Vesicular transport in Histoplasma capsulatum : an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol 10(8):1695–1710
48. Vallejo MC, Nakayasu ES, Longo LVG, Ganiko L, Lopes FG, Matsuo AL et al (2012) Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One 7(6):e39463
49. Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O et al (2007) Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6(1):48–59
50. Peres Da Silva R, Puccia R, Rodrigues ML, Oliveira DL, Joffe LS, César GV et al (2015) Extracellular vesicle-mediated export of fungal RNA. Sci Rep 5(1):7763
51. Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A (2009) Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155(12):3860–3867
52. Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR (2000) Urease as a virulence factor in experimental Cryptococcosis. Infect Immun 68(2):443–448
V. F. Santiago et al.
53. Huang SH, Wu CH, Chang YC, Kwon-Chung KJ, Brown RJ, Jong A (2012) Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One 7(11):e48570
54. Vargas G, Rocha JDB, Oliveira DL, Albuquerque PC, Frases S, Santos SS et al (2015) Compositional and immunobiological analyses of extracellular vesicles released by C andida albicans: extracellular vesicles from Candida albicans. Cell Microbiol 17(3):389–407
55. Wurdinger T, Gatson NN, Balaj L, Kaur B, Breakefeld XO, Pegtel DM (2012) Extracellular vesicles and their convergence with viral pathways. Adv Virol 2012:1–12
56. Lischnig A, Bergqvist M, Ochiya T, Lässer C (2022) Quantitative proteomics identifes proteins enriched in large and small extracellular vesicles. Mol Cell Proteomics 21(9):100273
57. Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE et al (2018) Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab 27(1):237–251.e4
58. Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A (2010) Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107(44):19002–19007
59. Lee JH, Choi CW, Lee T, Kim SI, Lee JC, Shin JH (2013) Transcription factor σB plays an important role in the production of extracellular membrane-derived vesicles in listeria monocytogenes. PLoS One 8(8):e73196
60. Jeon J, Mok HJ, Choi Y, Park SC, Jo H, Her J et al (2017) Proteomic analysis of extracellular vesicles derived from Propionibacterium acnes. Prot Clin Appl 11(1–2):1600040
61. Kim HY, Lim Y, An S, Choi B (2020) Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis. Mol Oral Microbiol 35(1):1–9
62. Wu Y, Chen W, Guo M, Tan Q, Zhou E, Deng J et al (2022) Metabolomics of extracellular vesicles: A future promise of multiple clinical applications. Int J Nanomedicine 17:6113–6129
63. Clos-Garcia M, Loizaga-Iriarte A, Zuñiga-Garcia P, Sánchez-Mosquera P, Rosa Cortazar A, González E et al (2018) Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J Extracell Vesicles 7(1):1470442
64. Iparraguirre L, Alberro A, Hansen TB, Castillo-Triviño T, Muñoz-Culla M, Otaegui D (2021) Profling of plasma extracellular vesicle transcriptome reveals that circRNAs are prevalent and differ between multiple sclerosis patients and healthy controls. Biomedicine 9(12):1850
65. Meningher T, Lerman G, Regev-Rudzki N, Gold D, Ben-Dov I, Sidi Y et al (2016) Schistosomal miRNAs isolated from extracellular vesicles in sera of infected patients; a new tool for diagnosis and follow-up of human schistosomiasis. J Infect Dis 215:jiw539
66. Macedo-da-Silva J, Santiago VF, Rosa-Fernandes L, Marinho CRF, Palmisano G (2021) Protein glycosylation in extracellular vesicles: structural characterization and biological functions. Mol Immunol 135:226–246
67. Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J et al (2019) Proteomic profling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol 2(1):325
68. Risha Y, Minic Z, Ghobadloo SM, Berezovski MV (2020) The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci Rep 10(1):13572
69. Batista BS, Eng WS, Pilobello KT, Hendricks-Muñoz KD, Mahal LK (2011) Identifcation of a conserved glycan signature for microvesicles. J Proteome Res 10(10):4624–4633
70. Martins ÁM, Ramos CC, Freitas D, Reis CA (2021) Glycosylation of cancer extracellular vesicles: capture strategies, functional roles and potential clinical applications. Cell 10(1):109
71. Borowski S, Tirado-Gonzalez I, Freitag N, Garcia MG, Barrientos G, Blois SM (2020) Altered glycosylation contributes to placental dysfunction upon early disruption of the NK cell-DC dynamics. Front Immunol 11:1316
72. Roca J, Rodriguez-Martinez H, Padilla L, Lucas X, Barranco I (2022) Extracellular vesicles in seminal fuid and effects on male reproduction. an overview in farm animals and pets. Anim Reprod Sci 246:106853
Another random document with no related content on Scribd:
Making T-Squares
B� J. B. Murphy
The making of a single article of any kind presents a distinct problem in itself, but the production of a large number of the same article must be done in a different way, if efficiency and uniformity in the product are desirable qualities. Taking, for instance, the making of a large number of T-squares, the material is not made up in the same manner as for one. Where a number of these instruments was required, they were made as follows, with no other Equipment than bench tools and a band saw. The squares were made of mahogany having both stock and blade edged with maple. The blades were fastened to the stock with five ³⁄₈-in. button-head screws.
Dimensions for a T-Square of Which a Number where to be Made in Duplicate
The material for the heads and the blades was glued up and finished to the sizes given at A and B. The material was cut to gauge lines on the band saw, the blades being a scant ¹⁄₈ in. in thickness, and the stocks, ³⁄₈ in. Two of each were cut from each prepared piece, first from one side and then from the other. They were then faced off on both sides, and two more pieces cut. With careful
cutting, six blades and six stocks were made from each piece. This left one side of each piece to be planed after sawing. The holes for the screws were drilled with a small hand drill.
Stock and Blade Material for Making the Parts, and the Jig for Assembling
For assembling, a jig was made by nailing a piece of stock, ³⁄₈ in. thick, to a straight drawing board. One end of the piece was planed straight and true before it was fastened in place. Stops were provided to locate the stock and hold the blade square with it. Wedges were used to keep both stock and blade against the stops while the screws were inserted. The wedges were not driven with a hammer, but pushed in firmly with the fingers.
¶Insert a scratch awl in common hard soap for hardening, and it will need no drawing after the plunge.
Paddling Your Own Canoe
by Stillman Taylor
PART II The Knack of Handling the Paddle
The knack of paddling a canoe as the majority of amateurs paddle is quickly learned, but to handle a paddle as the expert woodsman and the Indian wield it requires not a little practice and experience. In the birch-bark and canvas-covered craft, used by the guides of Maine and Canada, there are no seats, and the stern paddler kneels with his back against the rear thwart, while the bow paddler usually kneels with his back against the second thwart from the bow. However, the builders of modern canoes furnish a seat for each paddler, and while it is more comfortable to use them, it is better to take the kneeling position, which brings the weight lower in the canoe, as it is safer, and the paddler is better able to control his craft.
Again, the use of the seat makes it impossible for the paddler to put much of his body into the stroke, and he propels the craft largely through the muscles of his arms and shoulders. But when paddling from the knees they are spread out to brace the body firmly, and the back resting against the thwart enables the paddler to use his back and thigh muscles to drive his craft ahead with much less exertion and waste of energy than when sitting erect. Of course, the kneeling position is a source of discomfort at first, but after a little practice it will be easy to keep the position for several hours.
The stroke of the paddle is made with the body motionless, one hand grasping the shaft of the paddle and the other holding it a short distance above the blade. The paddle is now carried forward, dipped into the water in front, Fig. 1, A and B, and carried downward past the paddler’s body, Fig. 2, A and B, and the stroke is finished in the rear, Fig. 3, A and B. The paddle is recovered by turning the wrist so that the blade is turned edgewise as it is swung through the air to begin the second stroke. The swing of the paddle should be smooth from the beginning to the finish of the stroke, that is, free from undue jerkiness, but the long and slow sweep of the paddle is amateurish, pure and simple. The canoemen of the North woods do not use this stroke, but prefer a shorter stroke, quick recovery of the blade, and the “accent” of the stroke is much the same as that the expert axman uses when felling a tree. Therefore one’s energy should be concentrated upon the first part of the stroke, easing up quickly as soon as the blade is opposite the body, and finishing the stroke smartly as it is carried backward. The driving power of the paddle diminishes rapidly as the blade is carried backward, and if the full force of the stroke is continued with a long backward sweep, a way of most amateurs, much energy is wasted, since it adds nothing to the forward momentum of the canoe, but rather retards it, by forcing down the stern, and also causes the craft to drag more water after it. For a short afternoon paddle almost any kind of a stroke will suffice to drive the canoe ahead, but upon long trips, or when head winds and rough water are encountered, the quick, short stroke will save both time and muscle. The long sweeping stroke of the summer idler will probably average about 26 to 30 strokes per minute, while the North-woods guide and Indian will dip their blades almost twice as
often, a fair cruising average being about 45 strokes per minute. Short strokes, with the accent at the beginning of the stroke, will push the canoe faster and with less effort, and the paddler has better control over his craft.
F��. 1-A
F��. 2-A
The Stroke of the Paddle is Made with the Body Motionless, One Hand Grasping the Shaft of the Paddle and the Other Holding It a Short Distance above the Blade
The bow paddler usually does nothing more than keep up a regular stroke and watch for rocks and obstructions, but if he is a good hand at the paddle he can do much to steer the canoe. For example, take the usual bow stroke, made by dipping the paddle quite close and ahead of the canoe. The force of the stroke thus made forces the bow in the opposite direction, and the man in the stern will be called upon to use more energy in steering. In contrast to this faulty handling of the paddle, the experienced bowman will
F��. 3-A
F��. 1-B
F��. 2-B
F��. 3-B
begin his stroke by dipping his paddle well out from the bow and pulling it toward him at a greater angle, which keeps the canoe moving straight ahead without swinging to one side, and the energy of both paddlers is used in driving the craft steadily ahead. When paddling in tortuous and rocky streams it is the bow paddler’s duty to watch out for rocks and snags, and he should always be prepared to “draw” the canoe whenever it is necessary to clear an obstruction or assist the stern paddler to shoot the canoe around an abrupt bend. To do this is simple enough, since the bowman merely reaches his paddle out at arm’s length over the side and pulls the bow in that direction, while the stern paddler simply paddles straight ahead.
The stern paddler is the helmsman of the canoe, and while he can keep his craft on a straight course by trailing his paddle at the end of each stroke and using it like a rudder, it can only be done when a long, slow stroke is used. To save time and muscle, the experienced stern paddler always finishes his stroke with an outward push and a turn of the wrist, which offsets the swing of the bow and keeps the canoe on a straight course with much less effort than when trailing the paddle astern. Paddling from the stern is often considered difficult, but one soon masters it by a little experience.
Paddling in Open and White Water
Dangerous for the Novice, but Experts Win Races in This Position
When traveling in the open waters of large lakes where heavy winds and seas are commonly encountered, paddling should always be done on the knees, and the canoeist’s own experience and judgment must be called upon to meet constantly changing conditions of the wind and water. To cross a broad stretch of windswept water where whitecaps are running high, means a long, hard fight, and the stern paddler must be ever alert to ease the canoe through breaking crests and shift his balance whenever the necessity arises. When paddling in rough water, many canoeists make the mistake of driving their craft too hard, consequently the canoe will “pound” when it reaches the trough of the sea, and much unnecessary spray is sent flying aboard. Of course, in rough water
some spray is certain to come over the rail, but if the craft is well balanced by stowing the camp duffle so that the canoe floats on an even keel, it will, with proper handling, weather a pretty stiff gale.
When paddling against a heavy sea and wind, the regular stroke must be altered and modified, and the man in the stern may often balance his craft by holding his paddle in the water while a particular high comber sweeps by. Short and clean strokes are essential for handling the canoe in a heavy sea, and while the skillful canoeist can handle his craft in the trough of the sea when the need arises, it is hazardous for the novice to attempt it. In heavy weather, it is a good thing to keep one paddle in the water while the other is in the air, hence the experienced canoeist will—unconsciously perhaps— alternate or “split” strokes with his bowman.
To paddle, head on, into heavy seas is slow and tiresome work, and most canoeists find it easier to take the seas on the quarter, and the stern paddler must be ever on the lookout, nursing his craft over the big ones and often turning his bow into the largest combers. To be sure, the canoe will plunge more or less and yaw about, but the stern paddler merely checks this side spin, since he cannot prevent it entirely and drive his craft onward at the same time. When running before the wind, it is only necessary to keep the canoe balanced or “trimmed” to keep things dry. Here the bow paddler has the best of it; his sole duty is to keep paddling steadily ahead, while the balancing and handling of the canoe rests with the man in the stern. This point is well worth remembering, for to keep the canoe right side up when shooting rapids or when bucking heavy seas or a lake, the bow paddler must stick to his seat. He can, from his position in the bow, quickly note rocks or other obstructions and pass the word to his partner, who from his point of advantage can likewise better size up the effect of a high wave upon the craft. Probably more capsizes have resulted through the bowman’s mistaken efforts to balance the canoe by leaning to one side, or attempting to dodge the spray in rough-water paddling, than through the inability of the stern paddler to handle his craft.
Running a rapid stream in a canoe is replete with excitement, but it is likewise a dangerous bit of fun for the inexperienced canoeist who has not fully mastered the handling of his craft. When running rapids
where the water is fairly deep, the use of the paddle is the better way, providing both paddlers kneel in the bottom of the canoe. Unknown streams are naturally the most dangerous, although the veteran canoeist can “size” up the character of the stream at the first glance, that is, the trained eye of the woodsman will read the bottom of the river by the indications on the surface; depth of water, strength of current, swirl caused by submerged rocks, and many other details are evident to him. To the novice these signs mean but little. If the current flow is from three to four miles per hour, rocks or submerged logs, 4 or 5 in below the surface, will show very little, but in a 10mile or swifter current obstructions 12 or even 18 in. below the surface will throw up swirling ripples, and in a very rocky stream with much water flowing, rocks 3 or 4 ft. below will throw up waves of good size. Again, waves are caused by swift water striking the comparatively quiet water of a deep pool and the canoeist should be able to determine the difference between them in order to know what part of the stretch is dangerous and that which may be run in safety.
The waves in a rapid stream are altogether unlike the rolling waves of the open lake, in that the crests follow one another closely and the canoe cannot rise, but plunges through the curling crests. Rocks on the surface are easily avoided, as they split the current, and it is only necessary to steer the craft to one side to pass them safely. The one chief thing to do when shooting the rapids is to keep the canoe on an even keel and the bow into the current, and when it is necessary to change channels, the craft must be shot diagonally across the fast water. To keep the canoe under perfect control in white water, it must be propelled faster than the current flows, and the force of the stream must not strike one side of the bow with greater force than it exerts on the other side. When a canoe swings around in the current, the power of the paddles will be unable to cope with the more powerful current, which quickly swings it broadside and one is struggling in the water in the next instant.

The Double Blade Is Often Useful When Paddling Single-Handed on a Broad Reach of Wind-Swept Water
On a long canoe trip where much rapid water must be run, a “setting pole” should be reckoned a necessity. The pole may be a stout sapling, 10 ft. long, cut by the stream side, but an iron shoe, or spike, about 3 in. long, with an iron band or ferrule, should be carried in the kit, the iron spike being simply driven in the end of the green pole. The pole may be used to good advantage when running down shallow rapids, the man in the bow kneeling and using his paddle to help steer while his partner stands in the stern and wields the pole. When traveling up shallow and swift streams, the setting pole is absolutely necessary, and while at first it is an awkward implement to handle, a little practice will enable the canoeist to pole his craft with


























