

Foreword
Professor Philip O’Connell Executive Director
Westmead Institute for Medical Research
In September 1962, President John F Kennedy gave an address at Rice University where he said:
“We choose to go to the moon in this decade and do the other things, not because they are easy, but because they are hard, because that goal will serve to organize and measure the best of our energies and skills, because that challenge is one that we are willing to accept, one we are unwilling to postpone, and one which we intend to win […].”(1)
The address encapsulated much of what is good about humanity as it spoke to our best impulses, not our worst. Many of the sentiments of the address remain highly relevant today.
Firstly, Kennedy speaks of choice. The decision to take on this ambitious task was not mandated, or even essential, but chosen because of the benefits it would bring. Kennedy was appealing to a central component of the human condition, a desire to accept a challenge despite the risks and the odds.
The second point to emphasise is the audience he chose. He did not choose congress or a gathering of America’s elite, he chose a University, a place of learning and an audience of young men and women. He was appealing to the next generation to accept this monumental challenge.
The third point was the decision to take up the challenge precisely because it was hard. Rather than put this in a negative context, it was emphasised as a positive.
It was a challenge worthy of our time and effort. A challenge that required the buy-in of all of society, not just of government. This concept of undertaking the hard tasks, remains relevant to all of us today.
To all of the students that have committed to The Future Project at The King’s School and undertaken Science or Mathematics, you have not done this because they were easy subjects rather because they are hard, and worthy of your best efforts.
The sentiments that Kennedy articulated are just as important for you, as you embark upon the next stage of your life, as they were in 1962.
The challenges that your generation faces, such as climate change, energy transition, building better cities, or improving health, these should not be viewed as overwhelming but be seen as major goals that you choose to accept.
If you work as a team and bring all of society along with you, you can make the world a better place, through Science.
I congratulate all of you for taking up the challenge. It is the first step along the path to a life worth living. We were all put on this planet for a purpose. All that is required is that we rise to the challenges that we face.
1. Kennedy, J.F. 1962. Address at Rice University on the Nation’s Space Effort, September 12, 1962. Boston, Massachusetts: John F. Kennedy Presidential Library and Museum. https://www.jfklibrary.org/archives/other-resources/john-f-kennedy-speeches/rice-university-19620912
The
Contents
Foreword
Senior Interns—Quantal Bioscience
Exploring the potential of pigment-producing bacteria as sources of novel antimicrobials
Senior Interns—HydGene Renewables
Solutions for our Soil—Engineering Nitrogen Fixing Bacteria with Oxygen Sensing Capabilities
Junior Interns—Quantal Bioscience (Biology)
Assessing the Inhibition of Methylobacterium radiotolerans by Ethanol
The inhibitory effects of White Oil on the growth of Methylotrophs
Exploring methylotroph diversity by varying pH at isolation
Exploring methylotroph growth across varied salinity levels
The Captivating Quest for Methylotrophs
How Does Temperature Affect the Isolation of Methylotrophs from Leaves?
Junior Interns—Quantal Bioscience (Chemistry)
Microbial Pigments: Unleashing the Dye-namic Power of Microorganisms
Extracting Pigments from Methylotrophs with Oil
Ethanol improves pigment extraction from Methylotrophs
The Applications of Microbial Pigments
Junior Interns—Quantal Bioscience (Physics)
Faraday’s Legacy: Inducing More Power from a TENG
How Different Materials Affect the Voltage Output of a Triboelectric Nanogenerator
Contact TENGS vs Slide TENGS: Comparing Voltage Output
How the Surface Area of Electrodes Impacts the Electrical Output of a Laterally Sliding TENG
How Fast Can a TENG Go? – Frequency Versus Output
Serial Dilution: The Paradox of Adding TENGs in Series
TENGs in the MIST: The Challenge of Boosting Voltage using Series Connection
Junior Interns—The Westmead Institute for Medical Research
Conducting a Diagnostic Clinical Test to Assess the Risk of Ischaemic Heart Disease and Potential Myocardial Infarction
Robotics—Sydney Robotics Academy
Vex V5 Competition and Beyond
Robotics—Western Sydney University
Clara’s World—Bridging the Gap between High School and University

Exploring the potential of pigment-producing bacteria as sources of novel antimicrobials
Dinil Amarasinghe,1 Mehrub Chowdhury,1 Rushi Joshi,1 Krishna Li,1 Lauren Murray,1 Liana Niezabitowski,1 Shania Pimenta,1 Christian Pohl,1 Shaheer Abdul Rehman,1 Selvan Thillairajah,1 Harry Le Poer Trench,1 Dr. Belinda Chapman,2 and Dr. Michelle Bull2
INTRODUCTION
Antimicrobials are chemical compounds that slow or kill the spread of bacteria, viruses, fungi, etc, which are widely considered as threatening infections (1).
Antimicrobial resistance, also known as AMR, threatens the prevention and treatment of the infections previously mentioned due to the development AMR to combat modern medicine of antimicrobials (2).
AMR is a type of resistance developed in infections over time due to overuse of antibiotics and this causes medicine to no longer be able to respond to these infections which causes a risk of severe illness from simple infections due to this major resistance and causes epidemics and pandemics (2).
These potential risks from AMR infections have raised flags for a need for novel antimicrobials against the infections and if not recognized sooner, the growing proportions of infections will become untreatable.
Novel antimicrobials are a necessity to be able to treat these AMR infections and to discover these novel antimicrobials, there are various approaches. These approaches could vary from discovering new microorganisms producing a new type of antimicrobial to previously discovered microorganisms producing more effective antimicrobials (3).
Pigmented bacteria fall in the category of microorganisms producing bioactive products because of their pigment. Pigment production in bacteria are frequently isolated from natural environments such as soil or sand because their pigments as associated with resistance to UV light and other stresses in the environment whilst maintaining membrane stability and integrity (4).
The aim of this study was to isolate a range of pigmented bacteria from natural environments and investigate their potential to be sources of novel antimicrobial compounds.
METHODS
Isolation of pigmented bacteria from the environment
Samples of soil and leaves were collected from The King’s School grounds and sand was collected from Long Reef Beach intertidal rock pools.
Soil and sand samples were serially diluted in 0.1% peptone water and plated onto R2A agar (R2A), which is a specialized
and selective growth medium that supports the cultivation of a wide range of bacteria from environmental samples (5). Plates were incubated at 25ºC for 2 weeks.
Pink pigmented methylotrophs were targeted using selective agar containing methanol (5 mL/L v/v) as the sole carbon source (6).
Leaf impression plates were made by firmly pressing a leaf onto the agar surface to transfer bacteria from the leaf surface to the agar. Plates were incubated at 25ºC for 2 weeks.
Pigmented isolate identification
A selection of pigmented bacterial colonies from the different primary isolation plates were streaked for purification onto fresh agar. To identify the isolated bacteria, isolates were prepared for Sanger sequencing of the 16S rRNA gene.
An isolated colony of each isolate was suspended 1 mL sterile deionised water and centrifuged to pellet the cells. The washed cells were then resuspended in 5% Chelex (w/w) and DNA was extracted using a boil-lyse method.
The 16S rRNA gene was amplified by PCR using appropriate primers and controls. The amplified 16S rRNA gene PCR products were checked for correct size by gel electrophoresis against the positive control and a DNA size ladder.
The PCR products were then cleaned using a magnetic beadbased protocol and the DNA was quantified by fluorescence. The cleaned 16S rRNA gene PCR products were then Sanger sequenced.
Within 48 hours the FASTA files were received which contained the DNA sequence of the isolates. These sequences were then entered into a BLAST query and compared to the standard or 16S rRNA gene specific NCBI databases to identify the isolates to species level.
Lawn patch plate method
A lawn patch plate method was used to test for the production of antimicrobials from the pigmented isolates. Lawn plates of a test microorganism produce a thick, confluent growth of culture that is distributed uniformly across the growing medium’s surface and antibiotic sensitivity is tested by patching another microorganism onto the lawn.
A zone of growth inhibition around the patch can therefore be used to determine if antimicrobials are being produced. Senior Interns—Quantal Bioscience
The test microorganisms used were safe surrogates of the ESKAPE pathogens. The term “ESKAPE” is an acronym that stands for the following bacterial pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species
These pathogens are of particular concern in healthcare settings because they have developed resistance to multiple classes of antibiotics, making them difficult to treat.
We used the Risk Group 1 test bacteria as surrogates: Escherichia coli K12, Bacillus subtilis, Staphylococcus epidermidis, Lactobacillus plantarum, and Enterococcus faecium, and added S. cerevisiae as a representative a yeast.
For the lawn patch plate test, Plate count agar (PCA) and Tryptone Soy agar (TSA) were used and expected to support growth of the bacteria (surrogates and pigmented bacteria) and Tryptone Glucose Soy agar (TGYA) was used for the yeast S. cerevisiae
Lawn plates of the surrogates were made and cell suspensions of the pigmented isolates were inoculated with a loop onto the lawn plates in small patches (3 pigmented isolates per lawn plate). The plates were incubated for a duration of 1 week at 30ºC.
Pigment extraction from isolates
We extracted the pigments from each isolate to test for inhibitory effects of pigments directly against the surrogates. To extract the pigment contained within the cells, 1 mL of methanol was mixed in a microcentrifuge tube with a sample of cells.
Preliminary experiments with other solvents (ethanol and acetone) had shown that methanol was a suitable solvent.
Using sterile loops each culture was resuspended in the methanol and vortexed. The tubes were placed on rocker for 30 min at room temperature.
The cell suspensions were then centrifuged at 10,000 g for 10 min to removed cell debris and the supernatants were retained.
Microdilution Assay
To investigate the effect of the pigments on microbial inhibition, a range of pigment concentrations was prepared using a 1:2 ratio dilution method. Microtitre plate with 96-wells were used for the assay and pigments were diluted in a range of broths to support the growth of the test surrogates. Mueller-Hinton Broth (MHB) was used for testing the bacterial surrogates as it is known for its low concentration of inhibitors that could interfere with the growth of bacteria or the action of antibiotics (7).
The yeast surrogate was tested in Sabouraud Dextrose Broth (SDB). Triplicate wells were prepared for each pigment at each of 3 dilutions.
This setup allowed for a comprehensive assessment of how different concentrations of the pigment compound influenced surrogate growth.
Disk Diffusion Method
The Disk Diffusion by the Kirby-Bauer method was used to determine whether the ESKAPE surrogates could be killed or have its growth inhibited by different types of pigment extract. Lawn plates of the surrogate bacteria were made on MHA and yeast on Sabouraud Dextrose agar (SDA). Pigment extracts were pipetted (20 μL) onto sterile small paper disks which were dried for 5 min.
Afterwards, the paper disks with pigment extract on them were uniformly and evenly placed onto the lawn plates. Paper disks with 20 μL methanol were used as controls to ensure the validity of the results.
Disk diffusion plates were then checked after 48 hours to detect if there were any zones of inhibitions present around the disk that indicated that the pigment extract was inhibitory against that particular ESKAPE surrogate.
Effect of temperature and broth medium on growth and pigment production of S. marcescens
The growth of S. marcescens in different broths (Nutrient Broth [NB], MHB, and R2A Broth [R2AB]) and at different temperatures (25, 30, 25 and 40ºC) was tested. Sterile containers with 15 mL of each broth were inoculated with 100 μL of a S. marcescens cell suspension.
The containers were incubated for 1 week at each temperature. Growth was measured using a spectrophotometer (light transmittance at 600 nm) and visual observations were made for pigment production.
RESULTS AND DISCUSSION
Isolation of pigmented bacteria
Samples from locations within the Sydney region were cultured for the presence of pigmented bacteria. Surprisingly, only a very few bacterial colonies on the isolation plates from soil were pigmented. Many pink pigmented bacterial colonies of the same colony morphology were obtained from the leaf impression, but only one isolate was picked for further culturing.
The final cultured isolates came from The King’s School grounds (isolates 1, 2), a leaf impression (isolate 5) and three from Long Reef Beach sand (isolates 7, 8, 9). A range of pigment colours were produced by the isolates, all of which were bacteria. Table 1 shows the identity of the pigmented bacteria determined by DNA sequencing of their 16S rRNA gene and comparison to NCBI databases.
Table 1: Identity of pigmented bacteria isolated from different sources
1 Soil (TKS) Yellow
2 Soil (TKS) Red
5 Leaf (TKS) Pale pink
7 Beach sand (Long Reef) Orange
8 Beach sand (Long Reef) Pink
9 Beach sand (Long Reef) Yellow
Lawn patch plate method
Some methodological issues were identified for the lawn patch plate method after the agar plates had been incubated. L. plantarum did not grow to a confluent lawn on the PCA or TSA plates, possibly due to too low cell concentration at inoculation. Some lawns were also not swabbed onto the agar plates effectively, which made measuring any potential zones of inhibition difficult. No zones of inhibition were detected on plates where both the lawn and pigmented isolates had grown. New antimicrobial sensitivity media were used in further experiments.
Pigment extraction from isolates
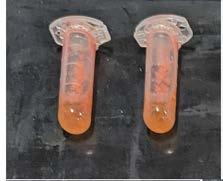
Pigments were able to be extracted from the bacterial isolates in methanol with 30 min incubation. The lowest apparent amount of pigment extraction was for M. extorquens (Figure 1).
Figure 1: Pigment extraction from each isolate using methanol as the extraction solvent (before centrifugation). D. maris (top left), M. extorquens (top middle), S. marcescens (top right), G. didemni (bottom left) and Q. aerophila (bottom right).



Disk Diffusion Method
The disk diffusion method was used to determine if the extracted pigments had an inhibitory effect on the ESKAPE surrogates through measurement the annular radius of any inhibition zones. The bigger the radius, the stronger the inhibitory effect of the pigment extract on the ESKAPE surrogate, indicating the pigmented isolate would have characteristics that might make it suitable as a candidate for antimicrobial production.
As seen in Figure 2, the paper disk with S. marcescens pigment extract created a zone of inhibition against B. subtilis (average annular radius of 4.5 mm) and S. epidermidis (average annular radius of 5.8 mm) thus indicating an inhibitory effect on thsee two ESKAPE surrogates. For the remaining agar plates, no pigment extract had an inhibitory effect on the ESKAPE surrogates.
Figure 2: Disk diffusion assay of extracted pigments against ESKAPE surrogates. Agar plates displaying S. marcescens (isolate 2) having an inhibitory effect on B. subtilis (top left) and S. epidermidis (bottom right).

Microdilution Assay
The microdilution assay results, as shown in Figure 3, indicated that M. extorquens and S. marcescens had a large inhibitive effect on most of the surrogates. M. extorquens was inhibitory to S. cerevisiae and B. subtilis S. marcescens was inhibitory against E. coli K12, S. epidermidis and B. subtilis. Inhibitory effects of both extracted pigments were observed at the highest dilution used in the assay. No growth of L. plantarum was observed in the MHB, so a new broth would need to be used for further investigations with this surrogate.
All the ESKAPE surrogates besides E. coli K12 are Grampositive bacteria. Despite Gram-positive bacteria having a thicker peptidoglycan layer, they are can be receptive to certain cell wall targeting antimicrobials due to the absence of the outer membrane (8). E. coli K12, B. subtilis and S. epidermidis were inhibited by S. marcescens while E. faecium, L. plantarum and S. cerevisiae (a yeast) were not inhibited. S. marcescens is a suitable candidate for antimicrobial production as it had an inhibitory effect on both B. subtilis and S. epidermidis
S. marcescens produces a red pigment called prodigiosin which has antimicrobial, antimalarial and antitumor properties. It has been demonstrated that prodigiosin inhibits growth of a wide range of Gram positive bacteria such as B. subtilis and Staphylococcus spp., as well as Gram negative bacteria including E. coli (9) affirming the results gained in the disk diffusion experiment.
M. extorquens produces carotenoid pigments which play a role in surviving oxidative stress (10). Ethyl acetate and chloroform extracts from other isolates of this species have been shown to have antimicrobial effects against B. subtilis, E. coli and Klebsiella oxytoca (11).
Figure 3a: Inhibition of ESKAPE surrogates by extracted pigments using a microdilution assay



Figure 3b: Inhibition of ESKAPE surrogates by extracted pigments using a microdilution assay

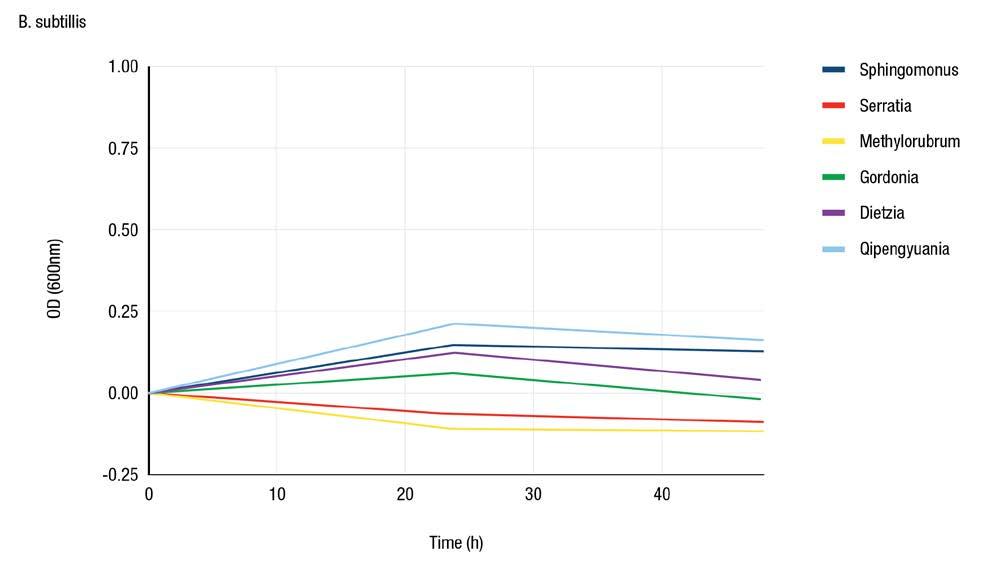
Figure 3c: Inhibition of ESKAPE surrogates by extracted pigments using a microdilution assay

The Effect of Temperature on growth and pigment production
Figure 4 shows how the optimal temperature for growth of S. marcescens varied with broth type. Growth was optimal in NB at 30ºC, in R2A broth at 35ºC, and in MHB at 2ºC. From visual observations of the different broths at different incubation
temperatures (Figure 5) we saw that pigment production was greatest at 25ºC and absent at 40ºC. At 25ºC, MHB resulted in the greatest pigment production.

Figure 4: Growth of S. marcescens in broth cultures incubated at different temperatures
Figure 3d: Inhibition of ESKAPE surrogates by extracted pigments using a microdilution assay
Figure 5: Observations of pigment production by S. marcescens in broths incubated at different temperaturesa

a For each image, cuvettes in order from left to right are NB, MHB, R2AB. S. marcescens broths incubated at 25ºC (top left), at 30ºC (bottom left), at 35ºC (top right) and at 40ºC (bottom right).
CONCLUSION
Through this series of experiments the potential of pigmented bacteria, or their extracted pigments, to have antimicrobial activity has been investigated. A range differently pigmented bacteria were isolated from diverse environments. These bacteria generally had limited antimicrobial activity, with the exception of M. extorquens and S. marcescens
Both of these bacteria have been previously reported to have antimicrobial properties, nevertheless, new isolates are of interest as they have the potential to have enhanced properties, including faster pigment production or pigment production under a broader range of conditions.
REFERENCES
1. Strohl WR. 2003. Antimicrobials, p. 336–355. In Microbial Diversity and Bioprospecting. John Wiley & Sons, Ltd.
2. Murray CJL. et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655.
3. Mantravadi PK. et al. 2019. The Quest for Novel Antimicrobial Compounds: Emerging Trends in Research, Development, and Technologies. 1. Antibiotics 8:8.
4. Ramesh C, et al. 2019. Multifaceted Applications of Microbial Pigments: Current Knowledge, Challenges and Future Directions for Public Health Implications. Microorganisms 7:186.
5. Reasoner DJ & Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7.
6. Delaney NF. et al. 2013. Development of an Optimized Medium, Strain and High-Throughput Culturing Methods for Methylobacterium extorquens. PLOS ONE 8:e62957.
7. Matuschek E. et al. 2014. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect 20:O255–O266.
8. Wright GD. 2010. Q&A: Antibiotic resistance: where does it come from and what can we do about it? BMC Biol 8:123.
9. Danevčič T. et al. 2016. Prodigiosin - A Multifaceted Escherichia coli Antimicrobial Agent. PLOS ONE 11:e0162412.
10. Rizk S. et al. 2021. Functional diversity of isoprenoid lipids in Methylobacterium extorquens PA1. Mol Microbiol 116:1064–1078.
11. Photolo M. et al. 2020. Antimicrobial and Antioxidant Properties of a Bacterial Endophyte, Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum Seeds. Int J Microbiol 2020:e9483670.
ACKNOWLEDGEMENTS
We would like to thank Dr. Michelle Bull and Dr. Belinda Chapman from Quantal Bioscience for their guidance during our TFP internship and for providing the resources. We would like to thank The King’s School for providing the opportunity of participating in The Future Project and to Loreto Normanhurst for their support of our involvement in the program.
Senior Interns—HydGene Renewables
Solutions for our Soil—Engineering Nitrogen Fixing Bacteria with Oxygen Sensing Capabilities
Ahmed Alani,1 Mayawen Devakumar,1 Anthony Foong,1 Ian Gao,1 Tom Hanley,1 Arney Kadaganchi,1 Asha Keshavarz,1 Ethan Low,1 Marcus Luck,1 Mylen Manivasahan,1 Austin Mclean,1 Skandaguru Murugabalaji,1 Daniel Payne,1 Ella Yumiao Peng,1 Arjun Raman,1
Tully Salmon,1 Erika San Diego,1 Taha Siddiqi,1 Wenxuan (Sara) Song,1 Tanush Veluppillai,1 Renqi Xu,1 Adrian Zhuang,1 Dr. Louise Brown,2 and Dr. Robert Willows2

INTRODUCTION
We are a team of 21 students from The King’s School, Loreto Normanhurst, and Tara Anglican School for Girls.
As interns with HydGene Renewables, we have been investigating the power of synthetic biology or ‘SynBio.’ SynBio is a suite of molecular tools that scientists can use to engineer biological organisms to do things they wouldn’t normally do in nature or perform tasks more efficiently.
This can include things like producing hydrogen from biowaste, which is helpful in tackling many of the world’s big problems, such as climate change.
In Term 1, the staff of HydGene Renewables upskilled us on a range of SynBio processes such as DNA extractions, restriction enzyme digests, and electrophoresis.
Once we had all had a chance to use a pipette and found where our strengths were, the team split into smaller groups, funnelling our brainpower and skill to where it could be best applied.
This included a wet lab team led by Ian Gao, a dry lab team led by Hugh, a business and marketing team led by Daniel, and a design and programming team led by Tom.
Our target goal with our project this year is to achieve a goldlevel award at the 2023 International Genetically Engineered Machine (iGEM) competition. iGEM attracts teams from around the world to share their innovative synthetic biology projects.
At The Future Project, we are proud to be developing a SynBio solution to address issues connected to Australia’s soil health.
Figure 1: The Future Project 2023 Senior Interns—HydGene Renewables
With our team’s project aptly named “SOILutions”, our team is investigating the complex nature of nitrogenase enzymes to produce bacteria that can fix nitrogen from the atmosphere to increase ammonia levels in the soil.
This solution is pioneering if the bacteria containing the nitrogenase enzymes can survive and function long-term in the soil. Survival is connected to the presence of oxygen in the soil, so we included an oxygen-sensing feature in our SynBio design to ensure the longevity of the bacteria.

If we can achieve our goals, then we can use this solution to improve soil quality without the need for fossil fuels.
While our project is actively contributing to the iGEM competition and remains in the development phase, we envision it as a promising prototype for a widespread soil enrichment initiative on a global scale. Our progress thus far is what we will share in this report.

PRELIMINARY EXPERIMENT
Over the course of three terms and two holiday workshop periods, we have been slowly piecing our project together by reading academic journals and interviewing experts.
We’ve also had team members working on the communication of our project, including producing promotional videos, Instagram posts, posters, and a website (also requirements of the iGEM competition).
To kickstart the experimental phase of our project, we first obtained a range of soil samples from around NSW to screen for the presence of diazotrophs. These unique bacteria are given this general name as they can fix nitrogen from the atmosphere to produce ammonia to deliver nitrogen to the plant.

The collected soil samples were first mixed with water and shaken for 30 minutes to extract the diazotrophs from the soil. The extracted bacteria were then inoculated into sample tubes containing a universal indicator.
If the colour of these solutions went more ‘bluish’ over time, we could assume ammonia was being produced as this is a basic compound. This simple test of colour change allowed us to deduce the presence of diazotrophs in the soil.
These tests screening the soil also provided us with crucial information on what healthy and unhealthy soil looks like and what we could potentially do to make it richer.
5: Semi-solid samples indicate the likely presence of diazotrophs by going blue

Figure 2: 2023 HydGene Renewables Senior Interns at work
Figure 3: Nitrogen cycle involving diazotrophs engineered by SOILutions
Figure 4: Soil samples from Parramatta all the way to the headwaters of the Hunter River
Figure
From a synthetic biology standpoint, we can then aim to engineer and manufacture our own diazotrophs, or the gene paths that they contain, to increase the ammonia content of the soil more effectively, making it healthier and nutrient-rich for plants.
Attempts to achieve this goal have been made before by researchers who inserted nitrogenase genes into E. coli bacteria.
However, they found this approach created a significant burden on the bacterial organisms for it to be constantly producing this enzyme.
Our project aim was to, therefore, develop a solution to this problem by introducing a pioneering oxygen-sensing system that could be used to turn on or off genes when required.
Getting bacteria to respond or do things under certain oxygen levels has also been done before. However, combining it with nitrogenase production, an anaerobic process that works best in low oxygen conditions, is a new solution application and one that can be used to address the soil health problem by ensuring that the organisms are only functional under conditions of low oxygen.
We are the first to try this approach. We hypothesise that the successful combination of oxygen sensing and nitrogenase function should increase the longevity of the engineered bacteria, allowing it to remain in the soil for longer, and continuously improve its quality or function.
OUR PROJECT
Our student ‘wet lab’ team read a range of scientific papers, particularly ones that looked at design strategies for a genetic-based oxygen regulation system.

Once a design was agreed upon, the next step was to get the oxygen regulator to work. We took what is known to be an oxygen-regulatory DNA-binding promotor in E. coli. It is called an FNR gene and can sense how much oxygen is present, allowing more RNA to be produced when the oxygen is at the right threshold and restricting it when the oxygen is not at a suitable level.
We then chose three promotors that are regulated by this regulator (FNR) to create a small genetic circuit, allowing us to control the production of a green fluorescent gene based on oxygen levels.

We synthesised the DNA for this plasmid in SnapGene and had it assembled and delivered in three parts. The parts were joined together using a synthetic biology process called a Golden Gate Assembly. These plasmids could then be replicated and inserted into E. Coli bacteria for testing.
The functional testing of our engineered bacteria showed that some colonies of bacteria did glow brightly under UV light Figure 9). However, the bacteria in the bottom left corner of this image did not.
Figure 9: Golden Gate selection — taking the right DNA based on its length
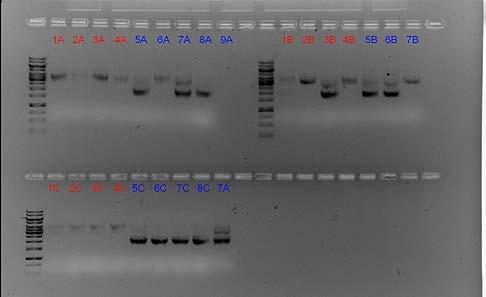
The colonies on agar plates that are glowing were kept at a low oxygen level, and the one that was not glowing was exposed to air. This was clear, convincing evidence that our oxygen-sensing system worked as planned!
We are continuing to do more testing to tune the response to oxygen. We then aim to replace the green fluorescent gene we have used for testing with a nitrogenase-producing gene cluster.
We look forward to continuing to work on this challenge during the upcoming term three holiday period. We are also excited to present our final project to the iGEM community in early November 2023.
Figure 6: 2023 Senior Interns collaborating with HydGene researchers during a term break intensive workshop
Figure 7: UV light image of bacteria colonies with this SynBio gene.

Assessing the Inhibition of Methylobacterium radiotolerans by Ethanol
Andrew Naeem,1 Austen Wong,1 James Wu,1 Johnson Zhuang,1 Dr. Michelle Bull,2 and Christine Chen3
1
The Future Project; 2 Quantal Bioscience; 3The King’s School
INTRODUCTION
Methylotrophs is a group of bacteria known for their capabilities in using single-carbon compounds such as methane and methanol as a carbon source (1). These bacteria are commonly found in a variety of environments, including soil, water, and even on plant surfaces (which excrete methanol).
Methylotrophs have been studied for their ability to degrade toxic compounds2 but the practicality of their use in reducing environmental contamination might be challenged by ethanol based disinfectants.
Ethanol is a commonly used antimicrobial agent due to its ability to disrupt cell membranes and denature proteins. It is widely used as a disinfectant and antiseptic usually at a 70% concentration in various settings, including medical, industrial, laboratory, and agriculture environments. Investigating the effect of ethanol on methylotrophs can provide insights into how the use of ethanol in these industries affect methylotroph growth and potential use.
OUR EXPERIMENT
The primary objective of this experiment was to assess the efficacy of ethanol at different concentrations in inhibiting the growth of Methylobacterium. We isolated a methylotroph from grass leaves, which we identified by DNA sequencing as Methylobacterium radiotolerans
By using an altered disc diffusion method, whereby paper discs were dipped into the aqueous ethanol solution, and placed onto a bacterial lawn on agar plates, we aimed to determine the inhibitory effect of ethanol on the bacterium’s growth and assess whether the size of the inhibition zone varies with different ethanol concentrations.

We conducted the experiment with 4 plates and 1 control plate (to ensure bacterial lawn growth in the absence of ethanol) and incubated it at 25°C for 2 weeks. The results were calculated based on the annular radius of the zones of inhibitions.
RESULTS
We expected, and confirmed with our results, that 70% ethanol concentrations would be the most inhibitory to M. radiotolerans, while lower concentrations would be less effective.
This demonstrates that the commonly used ethanol concentration used in disinfectants such as hand sanitisers or sprays would show some level of inhibition of the growth of M. radiotolerans, and thus affect its capability to use methane or degrade other compounds.
Figure 1: Ethanol sensitivity disc diffusion assay against M. radiotolerans

CONCLUSION
This study indicates that M. radiotolerans was most affected at higher concentrations of ethanol (i.e. standard of 70% in disinfectants).
Further studies would be needed to understand the underlying mechanisms and factors affecting inhibition.
REFERENCES
1. Chistoserdova, L., Kalyuzhnaya, M. G., & Lidstrom, M. E. (2009). Annual Review of Microbiology, 63, 477–499.
2. Dourado, M. N., Aparecida Camargo Neves, A., Santos, D. S., & Araújo, W. L. (2015). BioMed Res. Intern., 2015:909016
Figure 2: Inhibition of M. radiotolerans by ethanol
Junior Interns—Quantal Bioscience (Biology)
The inhibitory effects of White Oil on the growth of Methylotrophs
Ethan Armstrong,1 Preeth Kavin,1 James Napoli,1 Zichen Wang,1 Dr. Michelle Bull,2 and Christine Chen3
1The Future Project; 2Quantal
Bioscience;
3The King’s School
INTRODUCTION
Methylotrophs are a diverse and unique group of micro-organisms which can metabolise methylated compounds, a source of energy. These organisms are commonly found on plants and are prompting research regarding their possible contribution to reducing carbon emissions within our industrial world (1).
Within the 21st century modern landscape, agriculture stands at the forefront of creating a functional society. The rapidly universalised presence and application of organic pesticides and herbicides within an agricultural context has grown in consumer demand in the last decades.
This study investigates the inhibitory effects of white oil (a commonly used organic pesticide) on the growth of methylotrophic organisms.
METHODS
To conduct this experiment, varying amounts of white oil (Seasol brand) were mixed with diluent (0.6% peptone water) to produce incremental concentrations to obtain white oil to diluent ratios 1:16, 1:8, 1.4,1:2, 1:1 and 1:0.
Methylotroph isolate number 40 (isolated from a leaf press from Duranta erecta) was inoculated onto methylotroph agar plates as a lawn.
Sterile, blank filter paper discs were soaked into a single concentration of white oil and were placed in the middle of each third of the agar plates. The agar plates were incubated at 25˚C for 2 weeks.
RESULTS
The results demonstrate that white oil can inhibit the growth of methylotrophic organisms. Although there was no recorded inhibition of growth within the ratios of 1:16, 1:8, 1:4 and 1:2, it was evident that that the White Oil concentrations of 1:1 and 1:0 inhibited the growth (Figures 1 and 2).

Within these categories, 1:1 showed an average 2.67 mm radius of inhibited growth, with 1:0 showing an average 3.67 mm radius of inhibited growth, allowing it to be concluded that the white oil pesticide inhibits methylotrophic growth when utilised in high concentrations.
Seasol White Oil is a horticultural paraffinic oil designed to control pests on plants such as scale insects and whiteflies on tree, vine and vegetable crops.
Its properties, including a high paraffinic loading of 19.7g/L and effective leaf coverage, could potentially inhibit the growth of methylotrophic bacteria.
Compounds within the white oil act as a toxin, and as we observed, as the white oil concentration increases, so does the zone of inhibition.
Figure 1: White oil disc diffusion assay against a methylotroph (Isolate 40)

CONCLUSION
The ability of bacteria to rapidly repopulate has a crucial role in future experiments to enhance methylotrophic bacteria.
Further experiments such as exploring alternative pesticides options, inducing natural selection artificially through systematic testing, could determine the concentration of white oil required for methylotrophic bacteria to adapt and resist specific conditions.
REFERENCES
1. Chistoserdova, L., Kalyuzhnaya, M., & Lidstrom, M. (2009). Ann. Rev. Microbio., 63, 477–499.
2. Seasol (2022). https://www.seasol.com.au
Figure 2: Inhibitory Effects of White Oil on the Growth of a Methylotroph (Isolate 40)
Junior Interns—Quantal Bioscience (Biology)
Exploring methylotroph diversity by varying pH at isolation
INTRODUCTION
Methylotrophs as an antidote toward the excessive use of harmful chemical fertilisers has emerged in recent times as they play a significant role in the biogeochemical cycle in soil ecosystems, allowing for greater sustainable agriculture (1).
They ability to colonise different habitats of varying pH levels have therefore captivated the attention of scientists across the globe.
The aim of our experiment was to examine the diversity of methylotrophs from plants isolated at different pH levels.
METHODS
Samples of leaves from three plants (Gardenia, the Algerian Ivy and Southern Magnolia) were collected within The King’s School, which were then used in a leaf press, transferring existing methylotrophs from these plants onto selective methyltroph agar (MA) containing only methanol as a carbon source.
Each sample was pressed front and back separately onto MA at three different pH levels (6, 7, 8), leading to a total of eighteen plates. These agar plates were then incubated at 25ºC for two weeks.
Bacterial isolates from different leaves at the different pHs were further subcultured onto fresh MA agar. DNA from a colony from each isolate was extracted using a boil-lyse method.
A PCR for the 16S rRNA gene was performed and the amplicon was Sanger sequenced.
The 16s rRNA gene was targeted as it is present within all bacteria and can be used to identify methylotrophs to species level.
The obtained 16S rRNA gene sequence of each isolate was compared to the NCBI Nucleotide database using BLASTn to obtain an identity for each isolate.
RESULTS
Our results show that different methylotrophs could grow in environments of different pH levels.
For example, isolate 32, the back side of Gardenia leaf press incubated at pH 7 matched a known methylotroph isolated originally in a food factory in Japan (2), and has been shown to significantly increase plant yield in a biofuel crop (3).
Isolate 33 matched a known methylotroph named for its high radiation resistance (4).
Nicholas Chen,1 Oscar Hill,1 Shine Walker,1 Murphy Xi,1 Dr. Michelle Bull,2 and Christine Chen3
1The Future Project; 2Quantal Bioscience; 3The King’s School
Table: Identification of methylotrophs
CONCLUSION
We isolated a range of Methylobacterium species from the different plant leaves and showed that isolation agar at pH between 6 – 8 had minimal impact on methylotroph isolation.
Further experiments should be undertaken to show the complete diversity of methylotrophs on different plant species.
The ability of methylotrophs to grow at a range of pH levels make them well suited to use under a range of conditions to capitalise on their benefits which include the generation of ‘phosphorous, nitrogen, and carbon cycling [that] can help reduce global warming’ and as ‘biofertilizers for viable agriculture practices’ (1).
The additional property of methylotrophs, that of being able to grow on a single carbon source, makes this viable even in less-arable landscapes.
REFERENCES
1. Kumar, M. et al. (2016). World Journal of Micro. and Biotech. 32:120
2. Kato, Y. et al. (2008). Int. J Syst. Evol. Microbiol. 58:1134-41
3. de Aquino, G. et al. (2018). Industrial Crops and Products, 121, 277–281.
4. Ito, H., & Iizuka, H. (1971). Agric. & Biol. Chem., 35, 1566–1571.

Exploring methylotroph growth across varied salinity levels
Vikram Goel,1 Suhas Takkalapali,1 Shirane Bawa,1 Ishaan Devatha,1 Anav Pant,1 Riyaan Kamdar,1 Dr. Michelle Bull,2 and Christine Chen3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Methylotrophs have an ability to both degrade plastic materials (1, 2) as well as help make precursor compounds for biodegradable plastic via methane valorisation (3).
Plastic pollution in the oceans is a significant issue and solutions to rid the ocean of plastic of particular interest. Marine methylotrophs can accumulate on marine plastic pollution1
Our investigation measured the growth of a methylotroph isolate at different salinity concentrations. The purpose of this investigation is because the plastic within the ocean is associated with salt, and high salinity levels may disadvantage methylotrophic growth.
We tested the methylotrophs in 4 different salinity climates, 0.1%, 0.5%, 1%, 3% and 5%, trying to identify the limits of growth in a methylotroph.
METHODS
We isolated a methylotroph by performing a leaf press (from Malus pumila) onto methylotroph agar (MA) containing only methanol as the carbon source. An isolated colony was then restreaked onto fresh MA to obtain a pure culture.
We then tested for its ability to withstand different salinity levels by preparing a heavy cell suspension and swabbing it onto MA containing 5 different levels of salt (0.1%, 0.5%, 1%, 3%, and 5%).
RESULTS
We tested the ability of a methylotroph isolated from an apple leaf to be able to grow at different salinity levels (0.1%, 0.5%, 1%, 3%, and 5%).
The results, as seen in Figure 1, showed that there was an abundance of methylotroph growth when salinity levels were at 0.1% and 0.5%, meaning that methylotrophs will typically grow in environments with limited salt levels.
Methylotroph growth at 1% salt, had minimal growth and no growth was observed at 3%, and 5% salt.

CONCLUSION
The methylotroph isolated from a low-salt environment was not able to grow in environments with salinity levels greater than 1%.
The experiment was conducted in view of exploring the possibilities for plastic degradation in ocean conditions. Methylotrophs isolated from marine environments, or directly from ocean plastic, should have improved tolerance to high saline conditions.
An alternative is to perform further experiments to increase the salt tolerance of methylotrophs isolated from plants to better tolerate higher levels of salt associated with ocean plastic.
Finally, as methylotrophs are also associated with plastics in landfills and composts2, increasing the tolerance of methylotroph isolates to stress conditions, such as salt, may improve their biodegradation capability under other environmental conditions.
REFERENCES
1. Kelly, M. R., et al. (2022). Environ. Poll., 305, 119314.
2. Solano, G., et al. (2022). Environ. Chall., 7: 100500.
3. Zhang, M., et al. (2019). In: Methylotrophs and Methylotroph Communities. Caister Academic Press.
Figure 1: The results of each salinity levels
Junior Interns—Quantal Bioscience (Biology)
The Captivating Quest for Methylotrophs
Bryn Johnson,1 Rylan Prasad,1 Rohan Verma,1 Abe Wilson,1 Dr. Michelle Bull,2 and Christine Chen3 1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
The spotlight shines on two exceptional plants, Allium and Basil, as they reveal their hidden microbial companions in the captivating hunt for methylotrophic bacteria.
Methylotrophs play a significant role in the biogeochemical cycle throughout soil environments (ecosystems), fortifying plants and sustaining a developed agriculture1. Methylotroph isolation, used in biotechnological and scientific applications, is the process of gathering and acquiring pure cultures of microbes that can use one-carbon molecule as their sole energy source.
Methylotrophs can be isolated from methanol-rich samples, such as plant leaves2, and by culturing the microorganisms from the samples on selective agar plates that promote the growth of methylotrophs.
The aim of our experiment was to compare the number of methylotroph colonies isolated from different plants using two isolation temperatures.
METHYLOTROPH ISOLATION
In our experiment we aseptically collected leaves from two different plants (Basil and Allium) from The King’s School vegetable plot. Leaves were pressed onto methylotroph agar (MA), which contained methanol as the carbon source, using sanitized gloved hands to prevent contamination.
Three replicate agar plates for each plant were incubated at 25ºC and two replicate plates for each plant were incubated at 30ºC.
Plates were incubated for 2 weeks before observing for pink colonies, which were presumed to be methylotrophs2
RESULTS
The Basil leaves (Figure 1) had noticeably more methylotroph colonies and other bacterial colonies compared with the Allium leaves (Figure 2).
The methylotroph colonies from the Basil leaf impressions incubated at 30ºC looked to be larger and more numerous compared to colonies on MA plates incubated at 25ºC (Figure 1).

Less difference in methylotroph colony size due temperature of incubation was observed for the Allium leaves (Figure 2)
Figure 2: Impact of different temperatures on Allium

CONCLUSION
This investigation revealed that Basil leaves had a high number of methylotrophs, compared with Allium leaves.
Changing the temperature of the isolation plate incubation (25 vs 30ºC) had a lesser effect and different temperatures could be tested in future.
REFERENCES
1. Kumar, M., et al. (2016). World J. of Micro. & Biotech., 32: 120.
2. Yurimoto, H., et al. (2021). Microorganisms, 9: 809
3. Kumar, M., et al. (2016). World J. of Micro. & Biotech., 32: 120. Yurimoto, H., et al. (2021). Microorganisms, 9:
Figure 1: Impact of different temperatures on Basil
Junior
How Does Temperature Affect the Isolation of Methylotrophs from Leaves?
Shuwei Guo,1 Dayton Lo,1 Nie Tiger,1 Oliver Wu,1 William Yan,1 Dr. Michelle Bull,2 and Christine Chen3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Microorganisms, although microscopic, significantly shape ecosystems and global biogeochemical cycles. Methylotrophs stand out for their unique capacity to metabolize compounds few other organisms can (1).
This study investigated how methylotrophs could tolerate a range of temperatures, offering insights into broader microbial community responses to Earth’s changes.
Given Australia’s diverse climates, from hot summers to cold winters, our research delves into microbial influence on delicate ecosystem balance, aiming to uncover their concealed yet impactful role.
The aim of our experiment was to ascertain the temperatures in which methylotrophs would grow.
METHODS
Leaves from a Magnolia tree were picked, and the topside of each leaf was pressed onto individual methylotroph agar (MA) plates.
Plates were incubated at 8, 25, 30 or 39ºC for 2 weeks. Methylotroph colonies were presumed to be the pinkpigmented colonies (1).
RESULTS
Of the four sample temperatures tested, methylotroph colonies were observed at all temperatures, to varying rates of success (Figure 1). At the highest temperature, 40ºC, long term exposure to such high temperatures limited the number of the methylotroph colonies and moulds can be seen growing in the absence of methylotrophs.
At the cooler temperature of 30ºC, a temperature to simulate moderate environmental conditions, we saw a varying number of colonies between the replicate plates.
As shown in the Figure, one of the 30ºC replicate plates had a high number of methylotroph colonies almost covering the entire surface of the leaf impression.
The control variable was 25ºC, and performed the best out of our samples, methylotroph colonies covered the leaf impression for both samples and were visibly denser than all the other samples.
Figure: Methylotroph isolation plates incubated at 8ºC (top left), 25ºC (top right), 30ºC (lower left) and 40ºC (lower right).

Isolation of methylotrophs at 8ºC performed the worst, consisting of sparsely populated colonies, compared with at 25 or 30ºC.
CONCLUSION
Through this experiment, we found that methylotrophs, thrive in moderate conditions (25 and 30ºC) while they struggle to grow in extreme temperatures.
Nevertheless, some methylotrophs were able to be isolated under harsher temperatures (40 and 8ºC).
In our methylotroph isolation experiment emulating Australia’s seasonal temperature, and cultivating on agar plates, the response of methylotrophs to varying temperatures revealed insights into their preferred temperatures for growth.
This investigation sheds light on their adaptability and offers a simulated glimpse into distinct conditions in which they will thrive.
REFERENCE
1. Yurimoto, H., et al. (2021). Microorganisms, 9: 809.
Interns—Quantal Bioscience (Biology)

Junior Interns—Quantal Bioscience (Chemistry)
Microbial Pigments: Unleashing the Dye-namic Power of Microorganisms
Kevin Cho,1 Lucans Ivankovic,1 Rico Li,1 Josh Forbes,1 Dr. Belinda Chapman,2 and Renay Mannah3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Microbial pigments are pigments produced by microorganisms, including bacteria, fungi, and algae. These pigments are used in a variety of ways by microorganisms, for example, chlorophyll and phycobilins capture light energy for photosynthesis.
The study of microbial pigments offers insights into how bacteria adapt to various environments and has applications in industries like food, cosmetics, medicine, and environmental monitoring. In this article, we detail why microbial pigments are important chemicals and their relevant applications within the modern world.
Microbial pigments are valuable chemicals with diverse applications. They serve as sustainable alternatives to synthetic dyes in industries like food, cosmetics, and textiles.
Additionally, microbial pigments have bioactive properties, creating potential candidates for pharmaceutical and nutraceutical industries. They also play vital roles in environmental processes, acting as photoprotective agents and contributing to carbon fixation.
Therefore, studying microbial pigments can deepen our understanding of microbial physiology and ecology. Their significance lies in sustainable colouration, bioactive compound discovery and environmental applications (1).

Microbial pigments have a multitude of potential dyeing applications, ranging from food and beverages to textiles. They are used as natural food colourants that can make the appearance of food and drink look more pleasing.
Microbial pigments can be safer and more stable alternatives when compared to synthetic dyes which can majorly impact a person’s health.
Some microbial pigments possess antimicrobial properties and can inhibit the growth of certain pathogenic bacteria and spoilage micro-organisms. This helps to preserve food products and extend their shelf life.
Pigments like carotenoids and flavonoids have antioxidant properties that help to neutralise harmful free radicals found in the body.
Microbial pigments are also increasingly gaining traction as natural dyes for textiles. These pigments, sourced from a variety of microbes found in plants, water bodies, soils, insects, and animals, offer a sustainable option for dyeing fabrics.
In the 19th and 20th centuries, the demand for textiles surged. Thus, synthetic dyes emerged as a cheaper and more readily available alternative compared to natural dyes, albeit with environmental drawback (2).
However, the recent discovery of microbial pigments has revived interest in natural dyes, providing an eco-friendly substitute to unsustainable synthetic dyes.
REFERENCES
1. Ramesh, C., Vinithkumar, N. V., Kirubagaran, R., Venil, C. K., & Dufossé, L. (2019). Microorganisms, 7, 186.
2. Sen, T., Barrow, C. J., & Deshmukh, S. K. (2019). Frontiers in Nutrition, 6:7, doi: 10.3389/fnut.2019.00007.
Figure: Coloured pigments.
Junior Interns—Quantal Bioscience (Chemistry)
Extracting Pigments from Methylotrophs with Oil
Jack Q. Huang,1 George Huynh,1 Felix Jiang,1 Will Liu,1 Meng Xi Yang,1 Dr. Belinda Chapman,2 and Renay Mannah3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Microbial pigments are what creates colour in the environment around us, produced by microscopic organisms. Extracted microbial pigments are of interest to many industries as substitutes for synthetic dyes (1).
Chemicals extractions are the extractions that use hazardous chemicals to separate the pigment from plant or microbial cells. In some cases, chemical extraction can be highly effective at extracting pigments from specific organisms; methods including sub-zero freezing, using highly acidic or bases or the incorporation of technology such as HPLC.
A method that was successful in extracting Chlorophyll from leaves was dipping them in boiling water for 5-10 seconds then immediately immersing them in 80% acetone. This stops the formation of Chlorophyllide, the precursor chemical to Chlorophyll (2).
Green extraction is the removal of natural compounds and substances from raw materials without creating hazardous contaminants. This allows the use of alternative solvents, ensuring a safe extract.
Green extraction is useful for food and pharmaceutical industries as it allows for extractions of natural ingredients and flavours, contributing to the production of everyday necessities.
An example method is hydro-distillation, where plant material to go through the extraction process is put into a constricting container and heated with water as the solvent.
Whilst it is getting heated, the water boils, causing a mass production of steam which passes through the plant, vaporising the volatile compounds present within the plant. The volatile and pigmented compounds are collected at the end of a condenser (3).
COMPARING METHODS FOR MICROBIAL PIGMENT EXTRACTION INTO OILS
We tested the effect of different pre-treatments on the extraction of pigments from three pink-pigmented methylotroph isolates. The methylotrophs were extracted with or without a pretreatment such as bicarbonate soda, ethanol or heating before waiting for the pigment to be extracted into the oil for three days. The amount of pigment extracted was analysed using a spectrophotometer (Table 1, following).
Isolate M004, which had no pretreatment before soaking in coconut oil, had no colour change after the three-day period. The isolates M008 and M007 had the colour of the oil changed to a light pink. The high absorbance readings for M004 were due to interference from the coconut oil, not from any extracted pigment.
Table: Absorbance measurements of oil extracts from methylotrophs
CONCLUSION
The most probable reason on why isolate M004 failed was because it had no pretreatment, unlike M007 and M008 which were pretreated at high pH and with ethanol, and with heating (M007).
The different oils used for the extractions from all three isolates may also have affected pigment extraction.
REFERENCES
1. Padhan, B., Poddar, K., Sarkar, D., & Sarkar, A. (2021). Biotechnology Reports, 29, e00592.
2. Hu, X., Tanaka, A., & Tanaka, R. (2013). Plant Methods, 9, 19.
3. Ghosh, S., Sarkar, T., Das, A., & Chakraborty, R. (2022). LWT, 153, 112527.

Junior Interns—Quantal Bioscience (Chemistry)
Ethanol improves pigment extraction from Methylotrophs
Jack Honan,1 Jack Huang,1 Mercer Reed,1 Arthetya Srirangan,1 Dr. Belinda Chapman,2 and Renay Mannah3
1The
Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Methylotrophic bacterial communities are an important group of bacteria utilising reduced carbon compounds and play a significant role in plant growth promotion, crop yield and soil fertility for sustainable agriculture.
Methylotrophs are a diverse group of micro-organisms that are characterised by their ability to utilise compounds containing one carbon unit as their only source of carbon for energy for growth. They are crucial in Earth’s biogeochemical cycles, playing an intrinsic role in carbon and nitrogen cycles.
Recently they have contributed to many different studies including alternative biofertilizers and bioinoculants, fortifying plants and sustaining agriculture, and the carbon cycle between methane and CO2.
The aim for our experiment was to see the most effective way to extract pigments from methylotrophs.
METHOD
Six different methylotroph isolates were tested (isolates M007, M032, M004, M009, M012, M018). The variables for extracting the pigment that we tested included: pretreatment, type of solvent (lemon juice, ethanol, vegetable oil or coconut oil) and tuning solvent.
As a pretreatment we tested the application of heat to the methylotroph isolate cells once they had been resuspended in ethanol. Cells in ethanol were heated to 70˚C in a heating block for 2, 4 or 6 minutes and then further extracted in olive oil.
RESULTS
The use of ethanol on a heating plate of 70°C for 4 minutes effectively allowed us to extract pigment from the methylotrophs into olive oil.
Pink pigment was extracted from methylotroph isolates M007 and M018, which were the isolates treated at 70˚C for 4 minutes. Other isolates we tested, which were also treated at 70˚C, but only for at 2 or 6 minutes, however, did not result in pink pigment being released.
DISCUSSION
Our results indicated that treatment at 70˚C for 4 minutes with ethanol and using olive oil as a solvent proved to be the best method for extraction of pink pigments from some methylotroph isolates.
We compared our results to Laxmi et al who used a more laborious pigment extraction (1). Their extraction protocol used multiple organic solvent extraction steps, including chloroform, as well as an overnight incubation at -20˚C.
We noticed that Laxmi et al. had comparable amounts of pigments extracted to our experiment, despite their more time-consuming process. We therefore propose that the role of temperature in extracting pigments should be further investigated.
Throughout the process of this experiment, the most desirable conditions to extract methylotrophs effectively is to ensure the heating plate is used for not too short, but not too long; an ideal time of 4 minutes.
REFERENCE
1. Laxmi, J., & Mitra, B. (2012). Asian J Pharm Clin Res, 5, 52–57.
The Applications of Microbial Pigments
Christian Teo,1 Max Vukasovic,1 Andrew Wang,1 Daniel Xia,1 Harond Yu,1 Dr. Belinda Chapman,2 and Renay Mannah3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Microorganisms heavily rely on microbial pigments for various essential functions, including light absorption, defence against stressors like UV radiation, and support for nutrient assimilation. Furthermore, these pigments contribute to microbial community signalling and communication mechanisms (1).
Microbial pigments have drawn interest for their potential advantages for human life and businesses in addition to their importance in microbial ecosystems.
Vibrant colours may be produced naturally, which has various advantages over synthetic dyes and makes them a more sustainable and environmentally responsible option.
APPLICATIONS OF MICROBIAL PIGMENTS
Food and Beverage Industry
Microbial pigments are gaining popularity as natural food colourants due to their non-toxic nature and absence of harmful components. They are replacing synthetic colourants with health concerns and enhancing the visual appeal of diverse food products such as dairy, beverages, and confections (2).
Figure: Microbial pigments can be used as food colourants

Pharmaceutical and Cosmetics
In pharmaceuticals and cosmetics, microbial pigments are utilized as safe and non-allergenic colouring agents. They are employed to create visually appealing pills, tablets, and topical formulations without compromising safety or effectiveness (3).
Textile and Dyeing
Due to their eco-friendliness and capacity to give materials vibrant colours, microbial pigments have attracted a growing amount of interest from the textile industry.
By incorporating these pigments, the negative effects of traditional synthetic dyes on the environment and the health of dyers are lessened.
Photovoltaics
Certain microbial pigments, such as bacteriorhodopsin, exhibit photoelectric capabilities that make them promising for light-harvesting applications in photovoltaic devices.
Research is ongoing to harness their light-to-electricity conversion ability for affordable and sustainable solar energy systems (1).
Bio-Sensors
Microbial pigments have shown potential as bio-sensing agents due to their ability to change colour in response to specific environmental conditions or the presence of particular compounds.
These unique properties make them valuable for developing biosensing systems to detect pollutants, infections, and contaminants1
CONCLUSION
Microbial pigments are not only critical for microbial life but also have diverse applications across industries. Their natural origin, non-toxicity, and vibrant hues position them as attractive alternatives to synthetic pigments.
As we delve deeper into understanding these pigments properties and production processes, they hold the potential to drive innovation and sustainability across sectors, benefiting both the environment and human well-being.
REFERENCES
1. Ramesh, C., Vinithkumar, N. V., Kirubagaran, R., Venil, C. K., & Dufossé, L. (2019). Microorganisms, 7, 186.
2. Sen, T., Barrow, C. J., & Deshmukh, S. K. (2019). Front. Nutr. 6: doi.org/10.3389/fnut.2019.00007
3. Kiki, M. J. (2023). Cosmetics, 10(2):47

Junior Interns—Quantal Bioscience (Physics)
Faraday's Legacy: Inducing More Power from a TENG
Niko Hatzistamatiou,1 Frank Ji,1 Ryan Rahimi,1 Aanish Tewani,1 Derek Zhang,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
What is a TENG? A TENG, for reference, is a Triboelectric Nanogenerator². What’s a Triboelectric Nanogenerator? Well, imagine that you are in a desert, your phone is running out of battery and all you have are a couple cables, foil, and paper. What do you do?
Combine the foil and paper, connect the wires to your dying phone battery, and enjoy your triboelectric recharge created from your arm motion. This simple device creates a tiny amount of energy because electrons are moved from one material to the other on contact – known as the triboelectric effect.
The limitation of TENGs is that they produce a very shortduration spike of high voltage which is unusable by most electronic devices, which instead require a steady DC supply. While this can be partly overcome with rectification and filtering, the very short time duration of these spikes makes
Figure 1. Output Power vs inductance (number of coils)
it challenging to capture the energy created. Here at ‘For No Apparent Reason’ (FNAR), we explored including a Toroid inductor of variable inductance (measured in Henry).
The change in inductance was achieved by changing the number of turns of the inductor's coil and measuring the power output after rectification and filtering. The hypothesis by FNAR was that an increase in inductance would alter the voltage-time curve for the TENG. This occurs due to Faraday’s law¹: when a changing current passes through an inductor, a changing flux is created, inducing a back EMF. This back EMF acts to alter the shape of the voltage spike, both positive and negative peaks.
Modifying the spike in this way has the potential to increase the efficiency of the capture capacitor, by increasing the time for it to charge up, reducing the peak current which reduces energy loss (Ploss = I2R).

As has been shown in Figure 1, the number of coils on the Toroid inductor has a positive correlation with the power output of the TENG, with more coils on the Toroid inductor producing more power. The formula for this being P=(8.74*10^-3) * Number of coils + 2.21, with an R2 value > 0.8 suggesting a modestly significant result.
These results suggest that the inclusion of an inductor may improve the efficiency of a TENG when a DC output is required. We did not find a peak efficiency had occurred by N=80 turns (the limit of our apparatus), which forms a basis for further experimentation with increasingly high-value inductors.
Our data suggests that the inductor can increase power output by increasing the duration of the spike and facilitating the charging of the filtering capacitor at lower peak currents. Since, for most applications, a filtered DC power supply is required, this innovation increases the DC power output. Increasing the efficiency is a significant step towards TENG's being a viable alternative to batteries, particularly for remote sensing and body-worn biomedical applications.

REFERENCES
1. Marsden SR. LibreTexts Chemistry. Faraday's Law [Internet]. LibreTexts Chemistry. [cited date unknown]. Available from: https://chem.libretexts.org/Bookshelves/Analytical_ Chemistry/Supplemental_Modules_(Analytical_Chemistry)/ Electrochemistry/Faraday's_Law
2. Stilwell MD, Yao C, Vajko D, Jeffery K, Powell D, Wang X, Gillian-Daniel AL. Sparking a Movement: DIY triboelectricity experiments. The Science Teacher. 2021;88(3):30-36. Available from: https://www.jstor.org/stable/27135468

Junior
Interns—Quantal Bioscience (Physics)
How Different Materials Affect the Voltage Output of a Triboelectric Nanogenerator
Alex Cao,1 Eric Wang,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
A triboelectric nanogenerator (TENG) creates tiny amounts of electricity due the way that different substances hold onto their electrons, known as electron affinity¹. Materials can be classified on the Triboelectric series, where materials most distant on the series have the biggest difference in electron affinity and theoretically would produce the most electrical output.
In our experiment, we are testing how different materials will affect how much voltage the TENGS produce in a practical TENG.
Sliding contact TENGs generate a positive-going and negative-going voltage spike each cycle (left-right-left swipe) (Figure 1). From our experiments, these voltage spikes are not symmetrical with the negative going swipe appearing to consist of many superimposed peaks. The effect is greatly increased energy output.
Figure 1. Voltage vs time output for a sliding TENG as captured by a digital Oscilloscope

The triboelectric series ranks materials based on their tendency to gain or lose electrons when they encounter other materials. Materials at the opposite ends of the series have the strongest charge separation. When designing a sliding TENG, selecting materials from different positions on the triboelectric series can enhance charge transfer and energy generation.
The roughness of the material's surface can also impact the contact area and the mechanical interactions between the two materials during sliding2. A rougher surface might lead to increased contact and separation forces, which can enhance charge transfer. However, excessively rough surfaces might increase friction and wear, reducing the device's durability.
REFERENCES
1. Shawon, S. M. A. Z., Sun, A. X., Vega, V. S., Chowdhury, B. D., Tran, P., Carballo, Z. D., Tolentino, J. A., Li, J., Rafaqut, M. S., Danti, S., & Uddin, M. J. (2021). Piezo-tribo dual effect hybrid nanogenerators for health monitoring. Nano Energy, 82, 105691. https://doi.org/10.1016/j.nanoen.2020.105691
2. Kim, W.-G., Kim, D.-W., Tcho, I.-W., Kim, J.-K., Kim, M.-S., & Choi, Y.-K. (2021). Triboelectric Nanogenerator: Structure, Mechanism, and Applications. ACS Nano, 15(1), 258–287. https://doi.org/10.1021/acsnano.0c09803
Contact TENGS vs Slide TENGS: Comparing Voltage Output
Edward Huang,1 Stephan Shen,1 Yu Wang,1 Henry Zhang,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Triboelectric nanogenerators (TENG) hold the promise of generating electricity in remote applications from sources such as wave motion or the fluttering of leaves in a gentle breeze. Two promising designs are slide-mode TENGs and contact-separation mode TENGs, each of which exploits the differing electron affinity between materials to cause a net movement of charge.
Energy, generally provided through friction, liberates electrons which then transfer to the material with the higher electron affinity. This charge can be collected via an electrode and flow through an external circuit, discharging the material and allowing the cycle to repeat.3
The aim of this study was to compare the practical performance of Slide TENGs and Contact TENGs. This was achieved by moving a polyimide Disc from left to right to left at a constant speed for Slide-mode TENG and by pressing-releasing the disc at a constant speed for Contact-mode TENG1

The output of each TENG was rectified using a diode bridge and filtered with a 100 μF capacitor. A consistent operation of the TENG over 10 cycles was maintained and the DC output voltage measurement using a digital multimeter.

Figure 2. Voltage versus TENG Type
Figure 1. Slide-mode and Contact-mode TENGs
The results in Figure 2 showed that our Slide-mode TENGs produced of higher voltage on in comparison to a Contactmode TENG. This may be due to the constant sliding action creates more voltage due to the polyimide film being in constant direct contact with the paper and aluminium foil, while the contact based TENGs featured a separated and indirect contact with paper and aluminium foil which would result in the sliding TENG to be overall more effective.2
We additionally recorded TENG outputs with a digital oscilloscope. Sliding TENG generated both positive and negative spikes per cycle, whereas the contact TENG only produced a clear negative spike during each operation. The reduced efficiency in the "release" sub-cycle may have caused a charge buildup, hindering charge transfer in the subsequent "press" sub-cycle, likely contributing to the contact TENG's lower output. This further supports our above hypothesis that the separation of the collecting electrode during "release" and contact improvement during "press" may explain this phenomenon. To investigate further, we propose using conductive adhesive to maintain electrode contact.
The energy input to each TENG was not quantified in this experiment, as the amount of kinetic energy in practical applications usually exceeds the energy input required to liberate electrons.
Therefore, it is usually the practical ability of the TENG to harness movement (e.g., the tossing of a float in ocean ripples) to produce repeating cycles of charge difference between electrodes which is the limiting factor, rather than efficiency per se.
While sliding TENGs performed best in this study, contactmode TENGs have the potential to harvest energy from more diverse sources, like fluttering leaves and foot-shoe impacts. Further research to enhance their output could broaden their useful applications.
REFERENCES
1. Cheng, T., Shao, J. & Wang, Z.L. Triboelectric nanogenerators. Nat Rev Methods Primers 3, 39 (2023). https://doi.org/10.1038/s43586-023-00220-3
2. He, W., Liu, W., Chen, J. et al. Boosting output performance of sliding mode triboelectric nanogenerator by charge spaceaccumulation effect. Nat Commun 11, 4277 (2020). https:// doi.org/10.1038/s41467-020-18086-4
3. Wang, Z. L., Triboelectric Nanogenerator (TENG)—Sparking an Energy and Sensor Revolution. Adv. Energy Mater. 2020, 10, 2000137. https://doi.org/10.1002/aenm.202000137

Junior Interns—Quantal Bioscience (Physics)
How the Surface Area of Electrodes Impacts the Electrical Output of a Laterally Sliding TENG
Patrick Li,1 Daniel Yang,1 Irvin Zhang,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Triboelectric Nanogenerators (TENGs) are devices for transforming kinetic energy into electrical energy. They differ from traditional generators in that TENGs rely on triboelectric, not electromagnetic, phenomena.
Despite their tiny output, much research is being investing into this field as are superior to batteries for mobile power sources because they can produce power whenever the energy source continues and do not require expensive metals or chemicals1. For instance, oceanographic sensors can be powered by ocean ripples which never cease, or biomedical sensors powered by peristalsis continue without the risk of a chemical battery rupturing and harming the patient.
Most currently proposed applications of TENGs are for tiny devices and hence minimising the surface area of the electrodes is an important design consideration2. Therefore, we investigated relationship between the surface area (SA) of aluminium foil electrode on the electrical output of a TENG.
A TENG's raw output is short duration positive and negative spikes, so the output was rectified using a typical diode bridge (DIL 1A DB101) and filtered using a 47 µF electrolytic capacitor. This resulted in a smooth DC output suitable for measurement with a digital multimeter. A load resistor was used and the voltage across the load was measured, from which power could be inferred using an iteration of Ohm's law, P=VI.
A metronome was used to control the TENGs speed, and the reading was taken once the mechanical source could follow the metronome constantly and a steady-state voltage across the load resistor was maintained for 10 cycles. The gap between electrodes was controlled at 1 cm gap and the pressure was kept the same as far as possible between all TENGs.
As shown in Figure 1, the steadily increasing surface area of the electrode can be seen to have a positive correlation with power output. This demonstrates an approximately linear trend, suggesting that the amount of charge transferred to the electrodes is approximately proportional to their SA.

Figure: Surface Area of Electrode vs Electrical Output
An interesting minor deviation from the trend was consistently observed for the 20 cm2 TENG. A possible hypothesis might relate to the surface area of the sliding component which was approximately 15 cm2. It could be that the SA of the slider is a confounding factor and the similarity of the 10 and 20 cm2 results comes about because the area of the sliding collector could compensate for the smaller area of the electrode where is it below the SA of the slider. Further investigation to explore the most effective ratio of the SA of slider versus electrode may produce results more generalisable to the miniaturisation of TENGs.
A significant increase in the effective SA of each component of the TENG might be achieved by microstructures mimicking the microvilli found in intestinal walls, thus achieving the high SA in a tiny electrode. Such bio-inspired designs are starting to emerge in the literature3 and represent some interesting areas of future research.
REFERENCES
1. Cheng, T. H., Gao, Q., Wang, Z. L., Adv. Mater. Technol. 2019, 4, 1800588. https://doi.org/10.1002/admt.201800588
2. Yoon HJ, Kim DH, Seung W, Khan U, Kim TY, Kim T, Kim SW. 3D-printed biomimetic-villus structure with maximized surface area for triboelectric nanogenerator and dust filter. Nano Energy. 2019;63:103857. ISSN 2211-2855.
3. Li W, Pei Y, Zhang C, Kottapalli AGPK. Bioinspired designs and biomimetic applications of triboelectric nanogenerators. Nano Energy. 2021;84:105865. ISSN 2211-2855. doi:10.1016/j.nanoen.2021.105865.

Junior Interns—Quantal Bioscience (Physics)
How Fast Can a TENG Go? Frequency Versus Output
Caden Anbu,1 Arthur Bousquet,1 Gerald Chen,1 Manveer Dhaliwal,1 Krithish Roshan,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King's School
INTRODUCTION
When first hearing the word TENG, one may believe that it is somebody who has mispronounced the word ‘thing’. So, what is a TENG?
To elaborate in more depth, A TENG (Triboelectric Nanogenerator) converts mechanical energy into electrical energy through the triboelectric effect, which is achieved when materials come in contact and separate. It has gained popularity in many sectors such as renewable energy, sensing, and healthcare.
TENGs use materials distant from each other on the triboelectric scale and are useful for generating electricity from ubiquitous mechanical sources1 to power small devices, like our LED-illuminating TENG.
Some sources of mechanical energy can have very high frequency, such as the fluttering of leaves in the wind, which prompted us to explore how the output varies with frequency. Our sliding TENG's output was rectified using a diode bridge (1A, DIL) and filtered using a polarised capacitor. This resulted in a filtered output (measured using a digital multimeter) across the load resistor. From this, the power output can be inferred.
As observed in Figure 1, our tests discovered that when the TENG was slid at a higher frequency, more energy was produced. A linear relationship was demonstrated, with a higher frequency of sliding the TENG equating to a higher voltage produced, with an acceptable model fit (R2=0.84).

Figure: Voltage vs Frequency for a TENG
Two potential factors come into play. Firstly, a high frequency of operation results in more charge per unit time being captured, assuming that the net charge transfer per cycle is the same. If each swipe transfers C Coulombs of charge, then 10 Hertz should transfer twice the charge (10C) compared to 5 Hertz (5C) over 1 second.
The second factor is that when swiping at higher frequency across the same displacement, the relative velocity between the two surfaces necessarily increases. Velocity versus peak voltage output was tested separately using a digital oscilloscope to compare single swipes at varying velocities.
This also produced a positive correlation, meaning that a higher relative velocity produced a higher output.
These two factors imply that an increased frequency should deliver more discharges of charge per second, and each discharge should be higher (higher voltage) These two combined should produce a squared (parabolic) relationship between the frequency of operation and the output.
However, we observed a mostly linear trend. The data suggested a flattening at the highest frequency of operation –further challenging the prediction.
Further investigation of the velocity data captured by the digital oscilloscope revealed that while the peak voltage was higher, the duration of the spike was reduced, resulting the same net transfer of charge (the area under the curve. Thus, the assumption of equal charge-per-cycle holds and the linear trend as predicted by that assumption matches our measured data.
We propose a hypothesis that the slight flattening of the output at high velocities could be due to a reduced efficiency of the charge transfer. This may result in a rate limiting restriction on the output of a TENG at high frequencies of operation.
Thus, for effective high-speed TENGs, a lower resistance coupling of the electrode and the triboelectric material might be needed. We suggest ultra-thin triboelectric materials adhered to electrodes to reduce the surface-to-electrode resistance and increase charge transfer rate, may significantly increase the output of future high-frequency TENGs.
So no, a TENG is not the word ‘thing’ mispronounced—it has a huge potential waiting to be discovered.
REFERENCES
1. Weon-Guk Kim, Do-Wan Kim, Il-Woong Tcho, Jin-Ki Kim, Moon-Seok Kim, and Yang-Kyu Choi ACS Nano 2021 15 (1), 258-287 DOI: 10.1021/acsnano.0c09803

Junior Interns—Quantal Bioscience (Physics)
Serial Dilution: The Paradox of Adding TENGs in Series
Faris Hattouti-Rue,1 Luvin Idamegama,1 Rory Rengaswami,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
Triboelectric Nanogenerators (TENGs) are promising technologies2 for transforming mechanical energy into electrical energy. TENGs use triboelectric effect to transfer charge between two objects when they contact or slide against each other1
This transfer in charge is usually between high affinity and low affinity materials or positive and negative triboelectric materials. A limitation is that TENGS produce about 2 V and most electronic microcontrollers require 3.3 – 5.0 V. Our investigation was aimed increasing voltage output by connecting multiple TENG’s in series TENGs.
The series circuit was then rectified using a "bridge" of diodes which converted the AC output of the TENGs into DC. After that, filtering using an electrolytic capacitor ensured a smooth DC output which was recorded with a digital voltmeter. To synchronise the multiple TENGS, several operators attempted to slide their TENGs in unison using a metronome to keep a constant rate.
Our hypothesis for this investigation was that the more TENG’s we added the higher voltage output there would be. As shown in the table below our results show that the TENGs that were in series (No. 2 & 3) produce less voltage with averages of No.2 = 1.9475 V and No.3 = 1.9476 V. This is a surprising result, as according to Ohm's law voltages in series should add.

Figure 1: How the Number of TENGS in Series affects Voltage
Apart from the most striking feature of the decrease in output voltage with series TENGS, there was also significantly less variance in the N=2 and N=3 results, compared to the single TENG.
To investigate the surprising result of decreased voltage when in series, the raw output of a single TENG was recorded with a digital oscilloscope to capture the voltage versus time pattern.
This showed a positive going pulse as the slider was moved to the right and a negative going pulse as the slider was returned to the original position. A typical pulse duration was a few milliseconds.
From this observation we hypothesised that for TENGs in series, the coordination of the pulse had to be far more precise than was possible using our method. If the spikes of voltage did not exactly synchronise, then TENG 1 would peak while the others had zero output.
Outside of the moment of discharge, a TENG is a high resistance component and hence most of the spike from each TENG would be dissipated in the resistance of the other TENGs. This hypothesis is not fully supported by the data as it would predict a decreasing output with more in series (because resistances in series add), but instead the output appear to plateau for N=2, N=3. This curious result is worthy of further investigation.
We suggest two possible solutions for series TENGs in future research. Firstly, rectification and filtering could be done for each cell, and voltages would then add as expected as there would be no short-duration spike to synchronise.
Figure 2: Rotating Series Connected TENG

Secondly, a hyper-synchronisation of multiple cells may achieve perfectly simultaneous discharge which should then achieve the desired addition of the voltages.
We offer one potential solution through our invention of a rotating series connected TENG (Figure 2).
For this to work, each segment each "cell" would have to be hyper-matched to achieve simultaneous discharge.
Our results are a critical step in producing high voltage TENGs through series additional of multiple "cells".
REFERENCES
1. Triboelectric nanogenerator: Structure, mechanism, and applications ... (n.d.). https://pubs.acs.org/doi/10.1021/ acsnano.0c09803 Vivekananthan, V., Chandrasekhar, A., Alluri, N. R., Purusothaman, Y., Khandelwal, G., & Kim, S.-J. (2020, January 14).
2. Triboelectric nanogenerators: Design, fabrication, energy harvesting, and portable-wearable applications. IntechOpen. https://www.intechopen.com/chapters/70813
Junior Interns—Quantal Bioscience (Physics)
TENGs in the MIST: The Challenge of Boosting Voltage using Series Connection
Christopher Tiong,1 Owen Wang,1 Kevin Xiong,1 Dr. Roger Kennett,2 and Praneet Singh3
1The Future Project; 2Quantal Bioscience; 3The King’s School
INTRODUCTION
When was the last time that you used an electrical appliance? Electricity is all around in this modern time and reliance is only going up. While access to electrical energy is virtually limitless where a grid connection exists, mobile devices are still largely reliant on batteries.
For remote applications, batteries are problematic, requiring regular recharge or replacement which may be impractical1. In wearable technologies, button cells constitute a significant risk of harm if swallowed or if they leak their dangerous chemicals. It is for these applications that Triboelectric nano-generators (TENGs) are attracting great interest and investment.
Currently, though, they are limited by low voltage outputs and thus we attempted to explore connecting TENGs in series2 A series circuit is the common method to boost the voltage from battery powered circuits and thus seemed like a good starting point.
We realised from some initial observations using a digital oscilloscope, that the synchronisation of the TENGs in series would be critical as they deliver a very short duration "pulse" of electrical output which must occur at the same time for their voltage to add.
Therefore, we invented a device with multiple TENG components, driven by one sliding component, dubbed the MIST (Mechanically-synchronised Integrated Series Teng).
In Figure 1, triboelectricity is created by swiping a polyimide tape across then back on the piece of paper with Al foil, this is a simple TENG. Each additional TENG was wired in series with the rest to obtain an output of 1 – 4 series TENGs.
The output was bridge-rectified and filtered using a capacitor to create a smooth DC output. A digital multimeter was used to make the output recording once a consistent voltage was observed.

Figure 1. Mechanically-synchronised Integrated Series TENG (MIST) invention by the authors
A metronome app on a smartphone was used to ensure consistent frequency for all configurations.
We hypothesised that TENGs would act like chemical batteries where the voltage would sum by connecting them in series, in accordance with Kirchoff's law.
In Figure 2, there is no significant correlation between the number of series TENGs and the voltage output. This falsifies our hypothesis, as TENGs did not act like cells in a battery.
To explain this surprising result, we propose the hypothesis that despite our mechanical linking of each "cell" to ensure synchronisation of the movement, the moment of discharge or triboelectric charge transfer may still have a random element – much like how the exact moment lightning strikes is unpredictable.
Figure 2. Voltage versus number of TENGs
However, we are privy to other unpublished work by peers which showed a negative correlation between the number of cells and output – due to the high resistance of the other TENGs in the circuit. This research relied on human synchronisation.
Based on this evidence, we believe that our mechanical synchronisation method was at least partially successful. For this reason, we suggest that high tolerance manufacturing of MIST TENGs, could result in a viable, high-voltage output TENG array.
REFERENCES
1. Kim, D.W., Lee, J.H., Kim, J.K. et al. Material aspects of triboelectric energy generation and sensors. NPG Asia Mater 12, 6 (2020). https://doi.org/10.1038/s41427-019-0176-0
2. Niu S, Wang ZL. Theoretical systems of triboelectric nanogenerators. Nano Energy. 2015;14:161-192. ISSN 2211-2855. doi:10.1016/j.nanoen.2014.11.034

Junior Interns—The Westmead Institute for Medical Research
Conducting a Diagnostic Clinical Test to Assess the Risk of Ischaemic Heart Disease and Potential Myocardial Infarction
Finn McDonald1*, Udai Singh1*, Ryan Garg1, River Lusted1, John Foote1, Arhaan Trivedi1, Daniel Wang1, Edmund Dam1, Arjan Singh1, Wiley Gong1, Tharun Gowrinanthanan1, Rohan Jain1, Brandon Liu1, Sethin Ginige1, Ian Lee1, Hamza Arshad1, Jacob Khella1, Gaitan Njiomegnie2, Michael Nafisinia2,3
1

ABSTRACT
Introduction
Ischemic heart disease, affects around 1.64% of the global population, with an estimated 126 million individuals affected. Myocardial infarction (MI), commonly known as a ‘heart attack,’ is a severe consequence of ischemic heart disease, often driven by risk factors such as age, gender, high blood pressure, high cholesterol, smoking, obesity, diabetes, physical inactivity, and elevated low-density lipoprotein.
Methods
This study aimed to evaluate cholesterol and lactate dehydrogenase levels in two groups of patients, one with a predetermined healthy diet and the other with an unhealthy diet, all presenting with chest pain. Cholesterol levels were determined using spectrophotometric analysis. lactate dehydrogenase levels were measured with a clinical assay.
Results
Patients with unhealthy diets displayed significantly
higher cholesterol levels than those with healthy diets. Elevated lactate dehydrogenase levels were observed in all patients, irrespective of their diet, suggesting potential tissue damage. Patients’ conditions were categorised into eight scenarios based on their cholesterol and lactate dehydrogenase levels, chest pain, and electrocardiogram results, guiding appropriate management recommendations.
Conclusion
Elevated cholesterol and lactate dehydrogenase levels can be associated with a higher risk of MI. However, clinical symptoms, electrocardiogram findings, and specific biomarker tests, such as troponin, are essential for accurate diagnosis.
Early intervention and regular follow-up are crucial in addressing cardiovascular risk factors and monitoring the effectiveness of interventions. This study highlights the importance of follow-up assessments for individuals with abnormal cholesterol levels, heart biomarkers, and electrocardiogram results.
Proactive healthcare management can lead to early intervention, treatment, and monitoring, reducing the
The King’s School, North Parramatta, New South Wales, Sydney, Australia 2 Storr Liver Centre, Westmead Institute for Medical Research, Westmead Hospital and University of Sydney, Sydney, NSW, Australia 3 Discipline of Child & Adolescent Health, and Discipline of Genetic Medicine, Sydney Medical School, University of Sydney, Sydney, NSW, Australia
risk of cardiovascular diseases like heart attacks and strokes. Regular follow-up examinations are essential to track disease progression and treatment effectiveness.
INTRODUCTION
Globally, around one-third of deaths are attributed to cardiovascular disease. Ischaemic heart disease (IHD) is the most common type of cardiovascular illness, with an estimated 126 million people affected worldwide (Ojha N, Dhamoon. 2022).
This is equivalent to 1655 per 100,000 people, or 1.64% of the world’s population, and is expected to rise to 1845 per 100,000 people by 2030.
Furthermore, The Global Epidemiology of IHD Study (Ojha N, Dhamoon. 2022) found IHD to be responsible for 9 million deaths annually. Men are more commonly impacted by IHD than women, with incidence increasing with age from 40 years old.
Thus, it is evident that ischaemic heart disease is a significant problem in our society, and solutions are needed to address this growing problem.
A myocardial infarction (commonly known as a ‘heart attack’) is a severe consequence of IHD caused by a blockage in a coronary artery and resulting in a lack of oxygen and nutrients to cardiac tissue (Khan M, Kwiatkowski P, Rivera BK, Kuppusamy P et al. 2010).
There are several predisposing factors that increase the risk of myocardial infarction, including increasing age, male gender, high blood pressure, high blood cholesterol, smoking, obesity, diabetes, physical inactivity, and elevated low-density lipoprotein (LDL) (Hajar R. 2017).
Predisposing plaque build-up in the coronary arteries, known as atherosclerosis, is caused by LDL on the vessel wall. Then if the plaque ruptures or a blood clot forms, blood flow through the artery is obstructed. The prolonged lack of oxygen to cardiac muscle then causes it to die.
A myocardial infarction is, therefore, a major contributor to chronic heart disease and is one of the leading causes of mortality in the 21st Century.
Early detection and management of IHD and MI are essential in achieving the best outcomes for patients. Diagnosis relies on patient history detailing their symptoms, electrocardiogram (ECG) and different biomarkers. Initially, the patient notices physical symptoms such as chest pain, an ECG is used to determine if any abnormality may indicate cardiac damage, and finally biomarkers can confirm or rule out the presence of cardiac tissue death.
There are three main tests that are used to determine whether a myocardial infarction has occurred. These tests include Lactate Dehydrogenase (LDH), Creatinine Kinase (CK) and Troponin.
The LDH test was implemented in the 1960s for the detection of myocardial infarction. LDH is not specific for cardiac tissue, having five different sub-markers. It will also be elevated in other diseases such as lung disease, kidney disease, liver disease, skeletal muscle injury, as well as cancer, meningitis, encephalitis, and acute pancreatitis (Khan MA, et al. 2020).
In the 1970s the Creatinine Kinase test was developed. This test had three different sub-markers, one for the heart, one for the brain and one for skeletal muscle. CK was a more specific test than LDH, but still not perfect.
The troponin test is considered the gold standard for diagnosing myocardial infarction, as the troponin enzyme is only found in the heart. There are three troponin tests, with two being specific for the heart, Troponin I (TnI) and Troponin T (TnT). TnI is the better test, with higher diagnostic accuracy (Thupakula, S., et al. 2022).
MATERIALS AND METHODS:
1. Quantitative determination of total cholesterol levels in patient serum using spectrophotometric analysis
The reagents included:
• The Total Cholesterol reagents (Thermo fisher scientific, Cat number: TR13421)
• Total Cholesterol Standard Value, being 200 mg/dL (milligrams per decilitre)
• Patient Serum, which was diluted to 1:2 (collected 72hrs after MI symptoms)
• The Theoretical Control Value, being 200mg/dL
This was tested by pipetting 2.5 mL of total cholesterol working solution into 4 test tubes, labelled blank, standard, patient and control. Then 100μL of total cholesterol standard solution was added to the standard test tube, 100 μL of control solution was added to the control test tube and 100μL of patient serum to the test tube labelled patient.
Each tube was vortexed for around 5 minutes, and then incubated for 15 minutes at 37˚C. The absorbance values of each test tube were then measured in the spectrophotometer and recorded.
The total cholesterol levels were measured by dividing the absorbance value of the patient by the absorbance value of the standard and then multiplying by the standard value (200mg/dl).

As the dilution for the patient serum was 1:2 the result was then multiplied by 2.
2. Quantitative determination of lactate dehydrogenase (LDH) in patient serum using spectrophotometric analysis
The reagents included:
• LDH Reagents (Thermo fisher scientific, Cat number: TR20015)
• The patient serum (collected 72 hours after heart attack symptoms started to show)
Similarly, this was also tested by pipetting 2.5 mL of LDH reagent into the test tube. 100 μL of patient serum was then added to the test tube. The tube was vortexed for 5 minutes and then incubated for 15 minutes at 37˚C. The tube was then put into the spectrophotometer and the absorbance value was recorded.
LDH is measured in IU/L (international units per litre) and calculated according to the following equation:

Once the Factor was found, it was then multiplied by the Absorbance value to find the LDH activity. The results were then graphed.
RESULTS
Patients with healthy diets, all had cholesterol levels in the normal range (Graph 1).

In contrast, the patients with unhealthy diets had much higher cholesterol levels as demonstrated in Graph 2. Patient 4 had a cholesterol level, three times the normal level. This cholesterol level was nearly ten times higher than any of the patients with a healthy diet.

Graph 1. Cholesterol Levels in Patients with Healthy Diets

As both cohorts of patients, one with a healthy diet and the other with an unhealthy diet, exhibited concerning symptoms of a heart attack such as chest pain, nausea, and shortness of breath, we conducted LDH measurements, one of the cardiac enzyme markers, which was used to assess the potential elevation indicating damage to the heart muscle.
The normal range of LDH levels in the serum is typically between 114-240 IU/L, therefore, all eight patients had LDH levels that exceeded the upper limit of the normal range (Graphs 3 and 4).

Graph 3. LDH in Patients with Healthy Diets
Graph 2. Cholesterol Levels in Patients with Unhealthy Diets
Graph 4. LDH in Patients with Unhealthy Diets

These elevated LDH levels could indicate potential tissue damage or injury, including damage to the heart muscle. However, to confirm the possibility of a heart attack or other cardiac conditions, it would be appropriate to suggest further blood testing, including the measurement of CK (creatinine kinase) and troponin levels as well as electrocardiogram (ECG).
DISCUSSION
Elevated LDL is a known risk factor for ischaemic heart disease and myocardial infarction. Elevated cholesterol in this study correlated with diet, however, alone they are unable to determine whether a patient has had a myocardial infarction. Further assessment, including ECG and biomarkers, is needed in a patient with chest pain and elevated cholesterol.
Importantly, cholesterol levels can also vary with factors, such as age, gender, and individual health conditions. Therefore, it is always recommended to consult with a healthcare professional for a comprehensive analysis and interpretation of cholesterol levels.
LDH elevation can indicate myocardial infarction in the presence of clinical symptoms, however, it can also be elevated in other medical conditions. Therefore, using ECG and other markers such as Troponin would be important.
Using the cholesterol and LDH results obtained, the likelihood of myocardial infarction could be determined and the next important steps suggested. Patients in this study fell into eight different scenarios and management advice was devised -
Scenario 1: Patient with High Cholesterol, Normal LDH, No Chest Pain, Normal ECG
In this scenario, the patient only has a high cholesterol level, they should remain at home unless they start to show
symptoms such as chest pain. If symptoms do occur the patient is advised to call 000 or to check in with a doctor.
Scenario 2: Patient with High Cholesterol, Normal LDH, Chest Pain, Abnormal ECG
In this scenario, the patient should be taken to hospital as they are presenting with more specific signs of a Myocardial Infarction and should have further tests. It is important to note that LDH peaks after 48-72 hours, and this patient may see a rise in LDH while under observation.
Scenario 3: Patient with Normal Cholesterol, High LDH, No Chest Pain, Normal ECG
Here the patient should be tested if further symptoms and should be kept under observation. It is important to especially monitor the patient for chest pain and nausea as usually this is most indicative of a myocardial infarction and act according to the results received.
Scenario 4: Patient with Normal Cholesterol, High LDH, Chest Pain, Abnormal ECG
This patient should be taken to hospital immediately, as the patient is showing specific signs of an MI with chest pain and Abnormal ECG. Additionally, the LDH levels may have not peaked as of yet (as it peaks between 48-72 hrs).
Scenario 5: Patients with High Cholesterol and High LDH, with No Chest Pain and a Normal ECG
This patient may be at risk of having a silent heart attack, therefore the patient should be taken to hospital for further confirmatory tests such as troponin. High cholesterol and high LDH is not a good combination and it may be possible that the patient will suffer from a myocardial infarction soon (simply an assumption), if he is not already suffering from a silent MI).
Scenario 6: Patients with High Cholesterol,High LDH, Chest Pain, Abnormal ECG
This patient should be rushed to hospital as they are presenting with all signs of a Myocardial Infarction.
Scenario 7: Patients with Normal Cholesterol, Normal LDH, No Chest Pain, Normal ECG
The patient is recommended to stay home as they present with no symptoms or abnormal tests.
Scenario 8: Patient with Normal Cholesterol, Normal LDH, Chest Pain, Abnormal ECG
This patient is recommended to go to hospital as they may be having a myocardial infarction, further tests that are more accurate should be administered.
SUMMARY
In summary, proactive healthcare management plays an important role in identifying and addressing potential cardiovascular issues in individuals. In symptomatic individuals with elevated cholesterol levels, heart biomarkers, and abnormal ECG results, follow-up assessments become imperative.
Further, early identification of abnormalities provides an opportunity to implement lifestyle modifications, prescribe medication, and recommend appropriate interventions to reduce the risk of cardiovascular diseases such as heart attacks and strokes.
In addition, regular follow-up examinations enable healthcare providers to monitor the progression of abnormalities and assess the effectiveness of the prescribed interventions.
ACKNOWLEDGEMENTS
We would like to acknowledge The King’s School for implementing the Future Project Program, and we wish to express our special thanks to Ms. Shaz McAllister and Mrs. Samantha-Jane Odbert.
REFERENCES
1. Ojha N, Dhamoon AS. Myocardial Infarction. 2022 Aug 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan–. PMID: 30725761.
2. Khan M, Kwiatkowski P, Rivera BK, Kuppusamy P. Oxygen and oxygenation in stem-cell therapy for myocardial infarction. Life Sci. 2010 Aug 28;87(9-10):269-74. doi: 10.1016/j.lfs.2010.06.013. Epub 2010 Jun 28. PMID: 20600148; PMCID: PMC3114881.
3. Hajar R. Risk Factors for Coronary Artery Disease: Historical Perspectives. Heart Views. 2017 Jul-Sep;18(3):109-114. doi: 10.4103/HEARTVIEWS.HEARTVIEWS_106_17. PMID: 29184622; PMCID: PMC5686931.
4. Khan MA, Hashim MJ, Mustafa H, Baniyas MY, Al Suwaidi SKBM, AlKatheeri R, Alblooshi FMK, Almatrooshi MEAH, Alzaabi MEH, Al Darmaki RS, Lootah SNAH. Global Epidemiology of Ischemic Heart Disease: Results from the Global Burden of Disease Study. Cureus. 2020 Jul 23;12(7):e9349. doi: 10.7759/cureus.9349. PMID: 32742886; PMCID: PMC7384703.
5. Thupakula, S., Nimmala, S.S.R., Ravula, H. et al. Emerging biomarkers for the detection of cardiovascular diseases. Egypt Heart J 74, 77 (2022). https://doi.org/10.1186/ s43044-022-00317-2

Robotics—Sydney Robotics Academy
Sydney Robotics Academy—Vex V5 Competition and Beyond
Kevin Petrov, Leone Sun, Elliott Biggs, Alex Cai, Ulysses You, Azimkhon Mamatkhonov, Ethan W. Wang, Feifan Chen, Terrence Yong, Edmund Dam, Andrew Luu, Brandon Liu, Sethin Ginige, Jeffery Liu, Ian Lee, Sky Fan, Lucas Jin, Gavin Guo, Arav Pandit, James Zhang, Lawrence Wang, William Park, Johnny Shen, Lucas Guo, Tim Burykin, Angus Shui, Haochen Wang, Yu Wang, Henry Zhang, Patrick Li, Meng Xi Yang, Edward Huang, Stephan Shen, Abir Rahman, Nirupan Ananad, Hanson Hui, Rhetheesh Baheerathan, Aryan Haldankar, Tadhg Sheeran, Eduardo Robinson, Julian Dong, William Wang, Marcus Bosnic, Imesh Kariyawasam, Finn Jia, Ben Barraket, Lucas Lee, Isaac Bradford, Peter Xia, Sameep Oli, Jeremy Le, Dylan Ananad, James Mao, Hugo Yates, Stephen Zhong, Ben Veres


The Computing Studies department at The King’s School prides itself on nurturing student growth while embracing innovation.
It’s with this spirit we delve into the exciting world of The Future Project—a dynamic initiative aimed at embedding advanced computing science skills in students at The King’s School.
In 2023, students from Years 7 to 10 undertook an adventure of technological innovation through participation in the Vex V5 Robotics Competition, a platform that blends creativity, engineering, programming, project management, scientific acumen, and an aptitude for problem-solving.
The journey was challenging yet rewarding, filled with learning, camaraderie, and the thrill of competition.
Fuelling this ambitious initiative was the support of the Sydney Robotics Academy, whose invaluable expertise proved instrumental in propelling our students to new heights.
The academy’s professionals, equipped with their knowledge and industry experience, guided our young innovators on the path of robotics and computational thinking.
Jarod Li and his team at Sydney Robotics Academy have provided weekly support to the boys ensuring that they have access to the relevant expertise to augment their own knowledge.
The Vex V5 competition demanded our students push the boundaries of their knowledge, invoking their computational and engineering skills to build and program robots.
The contest prompted them to imagine, design, create, and iteratively improve their robotic models to compete in gamebased engineering challenges. This hands-on, minds-on process has been transformative, instilling in our students
a deeper understanding of technology’s workings and its potential applications in real-world scenarios.
Our students have excelled during these competition days, demonstrating immense aptitude and zeal. Through this journey, they’ve acquired key skills such as critical thinking, problem-solving, teamwork, and project management.
They’ve also learned the value of resilience and perseverance in the face of challenges—qualities that transcend the realm of robotics, permeating all aspects of life.
The success of The Future Project doesn’t merely lie in the competitive victories but more importantly, in the development of well-rounded, future-ready individuals who can navigate and contribute to an increasingly digital world.
This initiative forms the bedrock of our commitment to fostering an environment where our students are not mere consumers of technology, but active innovators and creators.
As we reflect on the accomplishments of the past year and gear up for the future, we extend our gratitude to the parents and the larger King’s School community for their unwavering support.
Together, we are shaping the engineers of tomorrow, paving the path for a future where technology and innovation are inherent parts of our societal fabric.
We are not merely preparing our students for a world that exists; we are equipping them to create the world that is yet to be.
The Future Project is a testament to that ethos, and we invite you all to join us in this exciting journey.


Clara’s World—Bridging the Gap between High School and University
Ashutosh Rai, David Allen, Edward Huang, James Wu, Joshua Wallach, Kevin Petrov, Krish Thakkar, Meng Xi Yang, Morgan Smith, Samuel Yin, Tom Tan, Tyler Mitchell, and Vedanth Trivedi
The framework used in the course is called Clara’s World which employs game-based learning and gamifi-cation approaches to motivate students to learn. Clara’s World is a fun and interactive platform that helps users learn programming in a visual and engaging way.
It is designed to teach Java programming through a series of exciting, real-world challenges. The platform is named after a small ladybug called Clara, who is the main character in the learning experience.
Clara starts with limited skills and learns programming by moving around and manipulating objects in her world. The creators of Clara’s World have been awarded several accolades, including the 2016 Educator of the Year in Asia Pacific Region by SEARCC and the 2015 ICT Higher Education Educator of the Year by Australian Computer Society 1.
The course was suitable for students who had some prior programming knowledge, and ran during Terms 2, 3 and 4. The students completed one project each fortnight and the projects included the following:
• Lesson 1, 2 - Basics of Java – Syntax
• Lesson 3, 4, 5 - Conditional Statements & Booleans –If statements – true, false
• Lesson 6, 7 - Loops and Creating Functions – While loops
• Lesson 8, 9, 10 - Variables – Printing – For loops
Students found the platform easy to use, and the challenges are designed to be both informative and entertaining. They enjoyed the fact that they could learn Java programming through a series of real-world challenges.
The platform is accessible on browser, tablet, or mobile device, which makes it easy to learn on the go. The best part about Clara’s World is that it offers a unique fun set of learning resources that are suitable for high school and university students.
Students from Years 8 to 11 embarked on a unique journey, delving into the world of Computer Science as they undertook the first unit of the Year 1 Computer Science degree from Western Sydney University. This wasn’t merely an introduction to the discipline; it was an immersion.
Guided by the sagacious mentorship of their tutor, David Coleman, students traversed the realms of Clara’s World, a specially designed online learning platform by the university.
The digital platform provided hands-on experience in coding with the Java programming language. But it was more than just lines of code; it was about unravelling the intricacies of software design and learning the principles that power the digital world.
Throughout Terms 2, 3, and 4, our students embraced rigorous academic content, translating it into tangible skills. They not only familiarised themselves with Java but also delved deep into core programming concepts, made available through an array of resources provided by the university.
Beyond the technical knowledge, the experience inculcated invaluable life skills. Students learned the art of organisation— how to manage time, prioritise tasks, and juggle between coursework, co-curricular activities, and personal commitments.
The program honed their computational thinking, empowering them to break down complex problems and design systematic solutions. They navigated challenges, sometimes individually and sometimes as a collective, refining their problem-solving abilities and learning the essence of teamwork.
The WSU - Clara’s World program has not only equipped our students with a head start into university-level computer science but has also instilled in them a confidence to face challenges head-on, to collaborate, innovate, and create.
As we close this chapter and anticipate the next, we remain grateful to Western Sydney University for this collaboration, particularly Anton Bogdanovych for putting the course together, to David Coleman for his unwavering support and teaching throughout, and to our diligent students, who continually inspire us with their passion and perseverance.
Robotics—Western Sydney University
Highlights
On 8 November 2023, The Future Project’s annual Graduation and Awards Evening celebrated the achievements of nearly 200 participants across Years 7 to 11 in eight programs with key industry partners.
In November 2023, the HydGene Renewables Senior Interns achieved a Gold Medal in the International Genetically Engineered Machine (iGEM) Competition for their Soilutions project submission. This the second consecutive year that King’s has received gold at iGEM, a remarkable achievement considering that this team was again the only Australian high school team to compete in this prestigious international competition.
The highly accomplished student robotics engineer teams in Years 7 to 10, in collaboration with Sydney Robotics Academy, also have achieved remarkable success in the Vex Tournament in 2023. Their achievements include the Excellence Award, the Amaze Award, the Think Award, and the Tournament Finalist Award in the NSW State Championships. The teams were also awarded the Innovate Award, the Tournament Champion Award, and the Robot Skills Champion Award in the qualifying tournament.










Acknowledgements
The King’s School established The Future Project to harness the transformational power of industry–school partnerships, with the aim to motivate and engage next-generation, globally minded scientists, engineers, inventors, critical thinkers, and pioneers. The Future Project creates collaborative opportunities with industry partners to build proficiency in innovative thinking, extension, and academic endeavour.
The Future Project is pleased to have collaborated with the following participating schools and industry partners in 2023 to deliver outstanding experiences and the opportunity for innovative discovery. Critical to program success in 2023 was the dedication and expertise of the industry partners, expert researchers, classroom teachers, and project supervisors.
The Future Project warmly congratulates all participating students, participating schools, industry partner personnel, and Schoolbased staff for their effort and collaboration.
PARTICIPATING SCHOOLS







