C R. Research Week. Special Edition.
COMMUNITY RESEARCH
AP Biology
Sugar and Saccharomyces cerevisiae 16
The Effect of SPF on the Mutation Rate of UV Mutated Saccharomyces cerevisiae 16
The Effects of Isobutanol on Saccharomyces cerevisiae 17
Yeast Fermentation on Sucrose, Fructose, and Glucose 17
Methods in Molecular Biology Research
Summary of Research Class 18
4DIU Protein 18
3HO4 Protein 18
2QRU Protein 19
4Q7Q Protein 19
2O14 Protein 19
3CBW Protein 20
Selection and Expression of Proteins with Computationally Predicted Structures 20
Smart Heart: Deep Learning-Based Analysis of 12-Lead Electrocardiograms for Cardiovascular Disease Risk Prediction 30
Studying Zooxanthellae to Protect Coral 31
SMART Team
The Dnase1L3 Flexible C-terminal Domain and Altered Charges in the Actin Recognition Site Result in Broad Substrate Diversity and Resistance to Actin Inhibition 32
Editor’s Note.
Welcome to the 2023 Research Week edition of the Pingry Community Research (PCR) Journal. We are excited to showcase Pingry’s top scientific talent, both in terms of research skills and knowledge of scientific concepts and discoveries.
The PCR journal provides students the opportunity to publish novel research. Through a written medium, students demonstrate their in-depth understanding of complex, collegiate-level scientific topics, and their applications in research at Pingry.
This special edition of PCR serves as a written complement to the in person poster presentations occurring throughout research week. Readers can preview abstracts, figures, and summaries reflecting the research conducted in advanced courses and extracurriculars, such as AP Biology, Independent Research Teams (IRT), and the Methods in Molecular Biology Research Class, among others.
Through the PCR journal, we hope to spark intellectual curiosity and promote scientific inquiry amongst the next generation of Pingry researchers.
Dive into the wonders of Pingry Research through this special edition of PCR: Pingry’s foremost journal of scientific research.
Evan Xie (V), Editor-in-Chief Annabelle Shilling (V), Editor-in-Chief Mirika Jambudi (VI), Former Editor-in-ChiefEditorial Staff.
Editors-in-Chief:
Evan Xie (V), current
Annabelle Shilling (V), current
Mirika Jambudi (VI), former
Copy Editors:
Leon Zhou (V)
Chelsea Peng (V)
Sophia Odunsi (V)
Julia Oussenko (IV)
Ella Tabish (IV)
Layout Editors:
Sarah Gu (V)
Leon Zhou (V)
Cartoon Editor:
Kain Wang (V)
Faculty Advisor: Mr. Maxwell

AP Biology.
Determining the Tun State Temperature Threshold of Tardigrade
by Thomas Goydan (V), Saniya Kamat (V), Grant Lardizabal (VI)The tardigrade is a semi-aquatic microorganism known for its ability to endure extreme conditions by undergoing cryptobiosis, withstanding temperatures ranging from near absolute zero to 151°C. The aim of our experiment is to determine the approximate temperature at which the tardigrade enters the tun state, a state of inactivity in which the tardigrade dries up and becomes dormant. To accomplish this, we used a pipette to collect tardigrades in small groups from a culture and then heated them in water using a hot plate. By observing the tardigrades at specific temperatures under a microscope, we found that the tardigrade enters the tun state at approximately 40°C. This research provides valuable insight into suspended animation, the temporary cessation of most vital functions without death, which operates in the same manner

as the tun state. Suspended animation could potentially provide doctors with more time to perform surgeries, effectively saving many lives.
Determining the Ability of Lemna minor to Remove Nitrogen From Water Sources
by Camille Collins (VI), Maddie Humphreys (VI), Stephanie Ticas (VI)Our project will provide information on Lemna minor’s ability to remove nitrogen from water sources, a potentially significant method to increase access to clean water. It is known that L. minor has nitrogen-removal abilities, so our project will provide further insight into its behavior in varying local environments. Specifically, we are measuring duckweed’s removal of nitrogen from three water sources, therefore
determining which source duckweed is most effective in. We gathered water samples from each source and created groups without duckweed, as a control, and groups with duckweed to measure the change in nitrogen concentration. Our results have yet to provide an answer, but they will be important in further understanding the nitrogen removal abilities and efficacy of L. minor
Determining the Mutagenicity of Propylene Glycol and Formaldehyde
by Clayton Annis (VI), Melania Rosales (VI), Grace Stowe (VI)Propylene glycol and formaldehyde are chemicals commonly found in vapes and e-cigarettes. Relatively new, these products have not endured extensive research on their potential long-term effect on humans. Our experiment is dedicated to discovering whether propylene glycol and formaldehyde are mutagenic. If so, they will pose a health risk for users of e-cigarettes and vapes. Formaldehyde is used in many types of glues, resins, and body embalming, and knowing whether or not these chemicals have mutagenic effects could lead to the usage of less harmful substitutes in the future. Initially, our experimental plan involved creating our own agar plates: one with adenine and one without adenine to simulate an environment where the implications of our mutagenic substances could be seen. Our results show that there is no statistically significant
data, as our p-value is .74393 and therefore, there is no convincing evidence that formaldehyde and propylene glycol have a mutagenic effect on the growth of yeast with and without adenine. However, solely based on the amount of yeast growth lost, formaldehyde yielded less of a loss than propylene glycol by 71.776 cm2. Our findings suggest that formaldehyde and propylene glycol do not have mutagenic effects. This does not necessarily mean that they are completely safe for human consumption as the data suggests that when these chemicals were added to the yeast, there was a significant decrease in colony growth compared to the control. Further experiments must be done in order to fully understand the effects of these substances on both yeast and humans.
Effect of Decreasing pH on Artemia franciscana Survivability
by Max Naseef (V), Leon Zhou (V)Ocean acidification, the decrease in pH of the Earth’s oceans as a result of the absorption of CO 2, is one of the foremost threats to marine ecosystems and by extension humans. It is therefore imperative to better understand the effects of ocean acidification on the survivability and response on a marine model organism like Artemia franciscana, brine shrimp. From this experiment, survivability thresholds can be determined and characterized before permanent damage is done to Earth’s marine ecosystems. 24 hour old A. franciscana was placed into three seawater solutions with varying pH (a control of 8 pH, and treatment groups of 7 pH and 6 pH).
The lethality of each solution at both 24 and 48 hours was then observed. Our 6 pH treatment was discovered to yield a statistically significant difference in survivability, meaning that if low enough, pH does affect the survivability of brine shrimp. This result provides insight into the future potential effects and sustainability of ocean acidification onto marine ecosystems and therefore human life.
Effect of Food Choice on Growth and Survival of Mealworms
by Ngozi Nnaeto (V), Monroe Russell (V), James Thomas (VI)Our project sought to investigate the effects of food choice on the growth and survival rate of Tenebrio molitor or mealworms. We assessed four diets, PVC plastic, PET plastic, styrofoam, vegetables, and a control of bran to investigate this research question. The mealworms were sorted into groups of ten, each with their respective diets. They were then bred for 25 days, and their growth and survival rates were recorded. However, despite constant upkeep, none of the mealworms on the vegetable diet survived. Several factors contributed to this, including excessive moisture and temperature. Therefore, the vegetable trial was excluded from our conclusion. The mealworms on the bran diet showed the highest growth, with a change in mass percentage of +400%, PVC with +200%, PETE with +190%, and styrofoam with +150%. A Chi-square analysis of the change in mass percentage yielded A X2 of
0.91, meaning we fail to reject the null hypothesis: Mealworms on the three diets will have the same survival rate. Bran also showed the highest survival rate of 90%, 80% of the PVC-dieted mealworms, 70% of the PETE-dieted mealworms, and 20% of the Styrofoam-dieted mealworms survived. Using the Chi-Square analysis, the survival data yielded an X2 value of 1.012. Thus, we failed to reject the null hypothesis: mealworms on the three diets will have the same mass. A larger sample size could confirm this result. This study highlights the importance of selecting appropriate mealworm diets for optimal growth and survival rates. Creating an efficient system using T. molitor for plastic reduction could provide a novel solution to one of the world’s most crucial problems.
Effect of Ultraviolet Light Exposure With Sun Protection Factors on Saccharomyces cerevisiae
by Jake Abdi (VI), McKenna Dwyer (V), Mary Claire Morgan (VI)The impacts of Ultraviolet (UV) radiation on the human body have received increased attention in previous years. The impacts consist of a series of illnesses, burns, and modifications in the genetic structures. The current study aimed to identify the effect of sunscreens on yeast (Saccharomyces cerevisiae) to prevent ultraviolet damage. Using 4 different SPFs (15, 30, 50, 75) on yeast grown on agar plates, we were able to compare the effects of UV light after five minutes. Furthermore, we used two controls, one under the UV with no sunscreen and one not put under the UV light. Our results showed
that the higher the SPF, the more yeast survived when compared to those with a lower SPF, or no sunscreen. This is greatly important given that it further enforces the significance of using sunscreen to protect your skin from the UV rays.
Effect of Varying Levels of Light Exposure on Lemna minor Growth
by Jaymin Bhat (VI), Tom Yanez (V), Asher Ziv (V)Lemna minor is a rapidly growing aquatic plant that is a model for studying basic plants. This study investigated the effect of varying light amounts on duckweed growth. An optimal growth technique for duckweed could be used to grow it at a controlled rate to reduce waste. The project involved using different benchmarks of shade covers and measuring the percentage of growth or decay over two week-long trials. Five microwells were used with eight individual plants per microwell with each grouping corresponding to a different number of sheets of a shade screen. Based on data from Trials 1 and 2, we could not determine a single leading amount of light that promoted growth. However, we were able to determine that both full light exposure and slight light filtering provide optimal growing conditions. In Trial 1, from Week 0 to Week 2, four shades of light caused 1.6 mm growth, while full light caused 0.7 mm growth. We concluded that despite our hypothesis, an increase in sunlight does not directly correlate with an increase
in plant surface area. Confounding variables such as evaporation and transpiration caused plants in each trial to die, resulting in data irregularities. In the future, temperature-controlled water and a standardized environment would be optimal for expedited duckweed growth.

Effects of Caffeine and Benadryl on Mealworms
by Grace Fernicola (V), Mia Libretti (V), Andy Overdeck (VI), Greta Pew (V)Our experiment will determine whether or not mealworms react to caffeine and Benadryl the same way that humans do. We sought to find the correlation between the movement of mealworms and the two drugs. A significant difference in movement would signify that mealworms to the drugs respond as humans do. We experimentally tried to answer our question by recording the movement of ten mealworms on camera for different caffeine and antihistamine concentrations. Our results were not very consistent. Our control group with no added drugs
to the mealworms’ food had the most movement. We expected the control to have a median amount of movement between the groups, but that did not occur. The caffeine groups also had less movement than the Benadryl groups, which was the opposite of what we expected. Our results show that caffeine and Benadryl don’t affect mealworms in the same way that they affect humans. We could have tried a different testing method to see if that would be more accurate.
Effects of pH on the Rate of Reaction of Norwegian Kveik Yeast
by Evan Wen (VI), Evan Xie (V)Alcohol fermentation is the process by which yeast, or Saccharomyces cerevisiae, converts glucose into ethanol, carbon dioxide, and ATP to fuel their cellular processes and produce societal essentials, including food, alcohol, and biofuels. Thus, it is critical to maximize the rate of reaction of yeast fermentation by optimizing the environment of the yeast. One promising yeast strain is Norwegian Kveik yeast, which has already been shown to ferment faster than traditional S. cerevisiae in the application of brewing. Since traditional S. cerevisiae have been shown to ferment fastest between pHs 4 and 6, it is likely that Norwegian Kveik yeast also has an optimal
pH range. Our research sought to find this optimal range by determining the rate of reaction of Norweigan Kveik yeast under different pH conditions. The reactions occurred in a solution of Norweigan Kveik yeast, sugar, water, and either hydrochloric acid or sodium hydroxide. The rate of reaction was determined by measuring the volume of carbon dioxide produced as a function of time using a balloon and water displacement. It was discovered that the rate of reaction of Norwegian Kveik yeast increases in more acidic and basic conditions, with acidic conditions yielding faster rates than basic conditions.
Effect of β-carotene on Protecting Saccharomyces cerevisiae Against Ultraviolet Inactivation
by Luke Evans (V), Trey Maultsby (V), Ally Smith (V)Saccharomyces cerevisiae, an extensively investigated yeast species, is a single-celled organism closely resembling human skin cells. Scientific literature has established that visible light of moderate intensity inhibits growth, respiration, protein synthesis, and membrane transport in yeast, also having a deleterious effect on membrane integrity. β-carotene, a color pigment in carrots, can pigment human skin post-ingestion to allow for better light absorption and prevent skin damage, as well as potential melanoma. This study aims to examine whether β-carotene is capable of protecting yeast from ultraviolet damage. Previous studies have reported that β-carotene is one of the microbial carotenoids produced by yeasts (BMC); however, the relationship between this antioxidant and UV exposure has only been investigated in humans, not in yeast. Lycopene, β-carotene, and lutein taken orally have also been found to aid in UV skin protection, but a 2014
study proved the combination just as effective as taking β-carotene by itself in preventing skin damage from UV radiation (NCBI). To further test our hypothesis that adding β-carotene to sucrose, baker’s yeast, and water solution will prevent the colony’s death due to UV exposure, YED plates with varying concentrations of β-carotene were exposed to UV rays. With a p value of .05, our chi-square of 2.769 is less than the critical value. Therefore, we fail to reject the null hypothesis because our data are not statistically significant. However, due to the limitations of our study, the protective effect of β-carotene on ultraviolet inactivation of S. cerevisiae remains to be seen.
Effects of Temperature on Ethanol Yield by Saccharomyces cerevisiae Fermentation

 by Jack Angell (VI), Charles Jiang (VI), Caleb Park (VI)
by Jack Angell (VI), Charles Jiang (VI), Caleb Park (VI)
Ethanol production by Saccharomyces cerevisiae is a critical process in the production of biofuels and alcoholic drinks. We examined the effect of different temperatures on ethanol yield by yeast production. We divided sugar water and yeast samples into three temperature groups (25 °C, 33 °C, and 42 °C). We measured the ethanol level daily until no changes were detected. Monitoring the
containers revealed that ethanol yields increase as the temperature does, until it reaches a specific temperature, where the efficacy decreases. Our work supports the importance of temperature on ethanol production. With this knowledge of how ethanol production responds to changes in climate and temperature, insights into fermentation optimization can be offered to several industries.
Nitrate Absorption and Effects on Growth in Lemna minor
by Jules Amorosi (V), Anika Sinha (V)Lemna minor, a small flowering aquatic plant, has garnered attention for its ability to absorb pollutants in polluted fresh or brackish water. In this study, we investigated the capacity of L. minor to absorb nitrate in different concentrations (0ppm, 5ppm, 10ppm, and 20ppm), and monitored the growth of twenty-four single L. minor plants over a period of fourteen or twenty-one days. Nitrate absorption was assessed using a nitrate electrode, while plant size was quantified using ImageJ. Our results reveal that L. minor exhibited greater nitrate absorption and growth in higher nitrate concentrations, underscoring its potential as a potent tool in mitigating water pollution. Our findings are significant in their implications for the realm of environmental pollution and water management.
Cartoon by Kain Wang (V)Optimizing Starch Concentration in Duckweed for Biofuel Applications
 by Brian Chin (V), Alex Massey (VI), Kyle Wang (V)
by Brian Chin (V), Alex Massey (VI), Kyle Wang (V)
Biofuels are part of the expanding sustainable energy industry that helps limit environmental issues by utilizing living matter like plants rather than crude oil, natural gas, and nuclear energy. The purpose of this research is to determine the pH level that optimizes starch concentrations in Lemna minor, or common duckweed, because starch can be fermented into useful biofuels like ethanol and butanol. Duckweed plants were grown under artificial light in solutions with pH levels 5 through 9 for two weeks. The starch was then extracted from each treatment group and combined with an iodine solution to measure concentration levels using a spectrophotometer. The experimental results showed that the duckweed plants grown in the
solution with a pH of 6 contained the greatest starch concentration (3.478 mg/mL) after the two-week growth period. This study will help inform the biofuel industry on the best growing conditions for L. minor in order to increase biofuel production and limit the use of fossil fuels.
Sugar and Saccharomyces cerevisiae
 by Kirsten Kamerkar (VI), Meher Khan (VI), Simone Powell (VI)
by Kirsten Kamerkar (VI), Meher Khan (VI), Simone Powell (VI)
Saccharomyces cerevisiae, or “brewer’s yeast,” is a vital ingredient for brewers and bakers. When combined with sugar in liquid, the yeast breaks the sugar down into energy via respiration. The more the yeast feeds on the sugar, the more influential it is as a raising agent in baking. The aim of this research is to determine which sugar is most effective at feeding yeast in a given time-
frame, thus having the fastest fermentation rate. There were four different sugars and a control, all used to measure sugar concentration from Day 0 to Day 7 across two trials. We ensured that each variable such as flask size, amount of sugar, temperature of water, and amount of yeast stayed controlled in order to obtain accurate results.
The Effect of SPF on the Mutation Rate of UV Mutated Saccharomyces cerevisiae
by Sophia Deeney (V), Noor Elassir (V), Siyara Kilcoyne (V)The lethal consequences of exposure to UV light in S. cerevisiae may be prevented by treating it with Sun Protection Factors (SPF). Evaluating the effect of SPF on the survival rate of S. cerevisiae can support SPF’s genetic improvement in preventing skin cancer in humans due to genetic similarities between humans and S. cerevisiae. In this study, S. cerevisiae was grown in a YED growth medium and its mutation rate at different concentrations of SPF with or without exposure to UV light was tested. A mathematical proportion was then utilized to compare the number of HA2 S. cerevisiae colonies mutated in each of the
test plates after the experiment to the initial number of colonies in the control plate. This ratio was employed to calculate the overall mutation rate of each of the colonies exposed to different concentrations of SPF with exposure or lack thereof to UV light and was plotted on a scatter plot. It was concluded that S. cerevisiae exposed to UV light in the absence of SPF exhibited a greater mutation rate than S. cerevisiae in UV light in the presence of SPF. It is evident that SPF not only protects the health of S. cerevisiae by preventing lethal UV light mutations, but also can be beneficial in prohibiting the development of skin cancer in humans.
The Effects of Isobutanol on Saccharomyces cerevisiae
by Sasha Gupta (V), Matteo Littman (V), Kyle Nicoll (VI)Our experiment focused on the effect of isobutanol on yeast fermentation of glucose into CO2. When yeast ferments glucose into CO2, it creates a highly efficient biofuel: ethanol. A byproduct of this reaction is isobutanol, an alcohol that sends starvation signals to the yeast and causes it to stop producing ethanol. Our experiment aimed to answer the question of at what concentration of isobutanol could yeast produce the most ethanol. To do this, we put yeast, glucose, water, and varying levels of isobutanol (10.834 mol/L) into Erlenmeyer flasks. We then put a balloon on each flask to catch the CO2 and ethanol. After a day or two, we tied the balloons and measured
their volume using water displacement. We initially hypothesized that a 0.5mL concentration of isobutanol (0.514 mol/L) solution would produce the largest amount of CO2. However, isobutanol produced the greatest amount in a significant difference between the amount of CO2 produced in the 1mL concentration, which had a molarity of 0.982mol/L. After increasing the concentration, the amount of CO2 produced became smaller. After using a 2mL concentration solution (a 1.808 mol/L), there would be no CO2 emitted. Therefore, using the solution with the 0.514 mol/L molarity is the most efficient way to produce large amounts of ethanol without killing the yeast.
Yeast Fermentation on Sucrose, Fructose, and Glucose
by Daniela Henriques (V), Sejal Patel (VI), Jessica Poprik (V)Saccharomyces cerevisiae, or Brewer’s yeast, is a unicellular fungus used to convert sugars into ethanol and carbon dioxide– a useful tool helpful to understand food processing. The purpose of this experiment was to determine what type of sugar would be most effectively fermented by SCOBY (symbiotic culture of bacteria and yeast) in the process of producing kombucha. Due to its simplest chemical structure, it was hypothesized that sucrose would be the most effectively fermented. To initiate the experiment, an equal amount of sucrose, glucose, and fructose were added to an equal volume of tea and SCOBY. Meanwhile, a Brix refractometer was used to record each respective sugar concentration at the date of making the kombucha and 16 and 44 days after fermentation. On the first day of fermentation, glucose, sucrose, and fructose all had
the same sugar content of 8%. 16 days after fermentation these levels remained the same. Lastly, 44 days after fermentation, fructose was still measured at 8%, sucrose at 7.8%, and glucose at 8.6% sugar content. Each level was expected to decrease over time, with the hypothesis sucrose would have the largest percent change. While the sucrose and fructose levels after 44 days of fermentation reflected more accurate results, the percent increase in glucose indicated an experimental error or a possible outcome unknown from past research, and therefore extra steps were taken to ensure accuracy amongst the equipment used. However, with the results procured, sucrose having had the greatest decrease in percent brix, supported the hypothesis that this type of sugar would most effectively be fermented by SCOBY.
Methods in Molecular Biology Research (I & II).
Summary of Research Class
Madeline Alfieri (V), Ethan Boroditsky (V), Charley Bush (VI), Julia Covello (VI), Alexa Drovetsky (VI), Isabella Fuentes (V), John Grissinger (VI), Charles Jiang (VI), Daniella Karnaugh (V), Adrian Kurylko (VI), Kellani Laidley (VI), Gordon Oatman (VI), Caroline Rewey (VI), Alex Rudnik (V), Ainsli Shah (VI), George Shavel (VI), Coco Simon (V), Javi Trujillo (V), Francesca Zarbin (V)
by Britney Alfieri (V),The BASIL (Biochemistry Authentic Scientific Inquiry) curriculum allows students in research classes to conduct novel research on specific proteins. These proteins have a known structure and researchers predict them to be enzymes, but they have not discovered their exact functions. Molecular Biology Research students attempt to determine the function of their assigned proteins, which takes multiple phases to complete. First, the students express the protein of interest in Escherichia coli cells. Second, students use computational tools to learn more about their POI (protein of interest) and to hypothesize a func-
4DIU Protein
by John Grissinger (VI), George Shavel (VI)We predict 4DIU is a carboxylesterase.
tion. Third, students must purify the protein in order to work with it more directly. After purification, students must develop an assay to confirm their hypothesized function. Finally, students will attempt to mutate their POI to deactivate it to determine the enzymatic function. This year, the two classes expressed their proteins successfully and predicted their functions with reasonable confidence. Currently, the classes are working on purifying their POIs from the E.coli expression cells. Once this process is complete, students will move on to designing activity assays to hopefully determine the function of their POIs.
3H04 Protein
by Daniela Karnaugh (V), Adrian Kurylko (VI), Gordan Oatman (VI), Francesca Zarbin V)3H04 is an enzyme that comes from bacterium Staphylococcus aureus. Using software such as the PDB, PyMol, Blast, Dali, and Moltimate, we hypothesized it to be a hormone-sensitive lipase-like carboxylesterase that breaks down carboxylic esters from substrates.


2QRU Protein
by Isabella Fuentes (V), Kellani Laidley (VI), Caroline Rewey (VI)2QRU is an enzyme with the predicted function of a carboxylesterase. We found this using the PDB, PyMol, Blast, Dali, and Moltimate.
4Q7Q Protein
by Ethan Boroditsky (V), Alexa Drovetsky (VI)4Q7Q is a protein that has putatively been characterized as an acetyl hydrolase using online database tools such as BLAST, DALI, and PyMol.
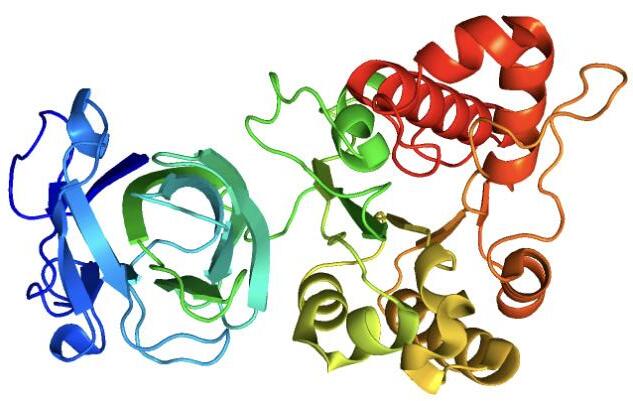


2O14 Protein
by Charles Jiang (VI), Ainsli Shah (VI)2O14 is an enzyme with an unknown function. Using PDB, PyMol, Blast, Dali, Moltimate, and other computational analyses, we predict it to be
a carboxylic ester hydrolase that hydrolyzes the carboxylic ester of a lipid into an alcohol and a carboxylic acid.
3CBW Protein
 by Maddie Humphreys (VI)
by Maddie Humphreys (VI)
3CBW’s known crystal structure, but unknown function makes it a critical protein of interest. Through previous studies including structural and computational analysis, it was predicted that 3CBW is most likely a beta mannananse–an enzyme that breaks down carbohydrates. The research conducted on 3CBW in wet lab and computational analysis for the rest of this year will intend to confirm that 3CBW is a beta-mannanase. These steps include expression and purification of 3CBW, site directed mutagenesis and expression/purification of mutants, and the pNPA & DNS activity assay. The outcomes of these experiments will provide a more contextual understanding of the 3CBW structure and pave directions
for future students to work on the 3CBW protein.
Selection and Expression of Proteins with Computationally Predicted Structures
by Adam Bauhs (VI)Every year, Methods in Molecular Biology Research I students research a specific protein pulled from a curated list of proteins with known structures but unknown functions. We use proteins with known structures so students can perform computational analysis on their proteins and determine future experimental directions. However, after years of running the class, many proteins on the list are no longer ideal for research. Some proteins have their functions, and some consistently express poorly in the lab, making study difficult. There is a need for a new generation of proteins for Research I students to study.
AlphaFold is an artificial intelligence program capable of highly accurate protein structure prediction. Nearly a million computationally created structures have been added to the RCSB Protein Data Bank (PDB), five times more than all previous experimentally derived structures. We sought to narrow this immense pool of novel structures into a list of proteins that we could
research in a laboratory setting. We accepted putative hydrolases as it allows Research I students to continue on a similar track for the first year. There are also many exclusion criteria based on maximizing the likelihood that the protein could express and catalyze in the lab.
We expressed ten proteins to gauge their expression levels in the lab. We found strong expression in eight of the proteins, moderate expression in one of the proteins, and weak expression in one of the proteins. This proof of concept has yielded eight new proteins for lab work in future years. Also, it indicates that our exclusion criteria successfully generate proteins suitable for lab work with a high success rate.
In the future, we plan to purify our proteins, record precise concentrations, and run assays. We also plan to use AlphaFold to generate more proteins for study as needed.
Creativity During Covid
by Maddie Humphreys (VI), Caroline Rewey (VI) Advisor: Dr. A. DickersonIt is impossible not to wonder how the Covid-19 pandemic will be represented in poetry, fiction, theater and film in the years to come. Tuberculosis, for example, figures prominently in Thomas Mann’s The Magic Mountain; Albert Camus, the author of The Plague, contracted TB at 17. This project seeks to discover how the pandemic has affected artists and writers’ creative process and material. Have these artists been able to write as never before, or has the pandemic disrupted their creative flow? Are they attempting to capture the
experience of the pandemic in their work, and if so, how? Has the pandemic altered their view of humanity and their experience of the world?
In addition to Pingry art teacher and painter Ms. Anne Ring, we interviewed the editor in chief of Pingry’s literary arts magazine, Calliope, Mirika Jambudi (VI), Pingry senior and acrylic painter Rachel Zhang (VI). We will be creating a visual magazine as the culminating project of our research.
Studying South Asian Representation in Media
by Jiya Desai (IV), Ananya Sanyal (V), Annika Shekdar (V), Aanvi Trivedi (III) Advisor: Mrs. U. AvilesSouth Asian representation in media of all forms has vastly improved over the years as a result of increased numbers of members of the South Asian community providing their insights into the South Asian experience. Our research team strives to uncover the experience of those people who straddle two cultures. How is their story told when told by someone from within the diaspora, and how is it told when told by someone looking in? To begin to discover this, we intend to go back to February 25, 1990, wherein Apu Nahasapeemapetilon made his first appearance in Season 1 Episode 8 of the Simpsons, entitled “The Telltale Head”. With the advent of this character came the first ever real represen-
tation of a South Asian character in mainstream media. As a relatively new initiative, we began this year diving into South Asian literature set as far back as the 70s. Through our initial literature review, we’ve pulled out what South Asian representation looks like and started a timeline tying South Asian literature to notable historical events in immigration history. We’ve begun examining the differences in South Asian characters depending on the voices behind them, and we hypothesize that South Asian literature and media written by South Asian authors are more impactful and authentic representations of the true South Asian experience.
IRT.
Accelerated Molecular Dynamics
by Jack Angell (VI), Eshaan Golchha (V), Ethan Liu (IV), Asher Matthias (VI), Alex Wong (IV), Hansen Zhang (V)Advisor: Dr. C.
The Accelerated Molecular Dynamics group is focused on developing numerical simulations for modeling motions of interacting particles and harmonic oscillator networks. Optimized N-body simulations based on propagation methods such as Verlet and 4th-order Runge-Kutta schemes are developed and compared on the MATLAB plat-
Chuform. Our simulations are used to study various forms of harmonic oscillators and their responses to external perturbations. Our research can be extended to explore more complex systems with different forms of forces such as crystal lattices and interacting particles under Yukawa potential.
Dnase1L3
by AJ Goel (IV), Mirika Jambudi (VI), Lauren Kim (VI), Lauren Poprik (V), Advisor: Dr. J. PousontDNA degradation is a critical post-cell death process driven by the Dnase1 family of endonucleases. A lack of DNA degradation can lead to the onset of human autoimmune diseases, including lupus and parakeratosis. One subfamily member, Dnase1L3, functions primarily in the liver. While studies have found that Dnase1L3 prevents the onset of Systemic Lupus Erythematosus (SLE), the exact catalytic mechanism for Dnase1L3 and its homologs remains unknown. Our project is focused on elucidating the structural features of Human Dnase1L3 and its associated animal homologs (Box Turtle and Owl Monkey), as no in vitro research has been conducted on the homologs. In the first year of our project, we have worked on transforming the Dnase1L3 homologs that we have and going through the procedures for autoinduction. For the future, our group will work on visualizing the 3D structures of Dnase1L3 and its homologs for comparison. Through our research, our group hopes to ultimately determine which variants can degrade DNA, and cross-reference structural features to make infer-
ences about the catalytic mechanism of Human Dnase1L3. Determining the catalytic mechanism of Dnase1L3 contributes to a growing body of research aimed at understanding the activity of Dnase1 endonucleases, and these insights could provide the basis for future targeted therapeutics in autoimmune disorders.

Drover: Drone-Rover Communication for Pathfinding
by Ayush Basu (VI), Ethan Chan (IV), Keira Chen (V), Connor Francis (IV), Charles Jiang (VI), Kotsen (IV), Nicholas Meng (V), Diego Pasini (VI) Advisor: Dr. M. JollyThe integration of an unmanned aerial vehicle (UAV, or drone) and an unmanned ground vehicle (UGV, or rover) into a single autonomous system (a drover) allows for unique abilities and visual perspectives that have many possible applications, such as a replacement for dangerous wildfire surveying techniques like hotspotting and cold trailing while providing real time data. This project is concerned with building a drover system cheaply and effectively and enabling it to cooperate and pathfind in unknown environments.


We propose a cooperative pathfinding protocol called Long-term Direction, Short-term Correction (LDSC), where the drone’s global view allows it to dictate paths (long-term) for the rover, and the rover uses LIDAR to identify smaller objects in its path and dodges them (short-term). The drone’s path-planning ability involves turning images into node maps and applying the A* search algorithm. In developing the drone end of the LDSC algorithm, we found
that weighing the nodes by color was the most effective for pre-planning paths. Following the calculation of the path from the drone image, the rover uses an algorithm involving pre-calculated proportions and the Vincenty formula to convert the A* pixel path into a GPS path. We developed a wall-following algorithm to help the rover avoid obstacles invisible to the drone.
In order to ensure scalability, we used Apache Kafka stream processing to communicate between vehicles in real time. Testing of the pathfinding system with satellite imagery yielded consistent, centimeter-accurate results necessary for high-precision autonomous navigation when investigating hotspots in dynamic environments. Ultimately, the LDSC algorithm will be able to generate these paths on continuous footage, translate them into instructions for the rover, and allow the rover to efficiently alter these instructions in the short term.
Expression and Analysis of Fluorescent Fish Proteins In Vitro
by Ethan Boroditsky (V), Morgan McDonald (VI), Nataly Ruiz (IV), Ziv Shah (IV), Annika Shekdar (V), Rachel Zhang (VI), Leon Zhou (V) Advisor: Dr. D. FriedThere is an ever-growing need for improved fluorescent tags in the medical field. The discovery of the Japanese freshwater fluorescent eel (Anguilla japonica) in 2013 is evidence of the first fluorescent protein to be identified in vertebrates; one step closer to more efficient fluorescent applications. Since then, several fluorescent marine organisms have been identified, including lizardfish, scorpionfish, seahorses, and even turtles. We currently aim to purify red fluorescent fatty-acid binding
proteins from scorpionfish (Scorpaenidae). To fluoresce, these proteins require the cofactor bilirubin, but currently, researchers have not been able to bind the bilirubin in vitro and achieve fluorescence. The scorpionfish protein has been expressed to high purity and the next step is attempting larger-scale expression and purification of our protein, determining how to bind bilirubin to the protein, and attempting to induce fluorescence.
Extended Reality Research at Pingry
by Sia Ghatak (IV), Parth Patel (IV), Ananya Sanyal (V), Rohan Variankaval (V), Kain Wang (V) Advisors: Dr. D. Mirliss, Mr. A. SteinbergThrough our literature review, we identified a few common challenges with educating people about climate change. Most problematically, many of the prominent climate change issues, such as air pollution or ocean acidification, are invisible to the naked eye. In a physical sense, the subjects’ related particles are extremely small; more personally, these climate issues also either only affect a subset of the population, or have impacts far enough in the future to stir indifference. On this note, those who have the largest impact on climate change often do not face the consequences, creating a lack of understanding between their actions and the true impact said actions can have on the environment. The latter change is especially prevalent with our target demographic in sharing this information with the high school population. Presently, people, especially students, spend less and less time in nature, leading to lesser concern for the environment, and thus indifference to the possible difference their actions could bring. After discerning the subject of our simulation, we held a discussion with VR specialist Anna Carolina Muller Queiroz, an author of the paper “Virtual reality as a promising tool to promote climate change awareness,” to further our understanding of virtual reality’s use in
promoting climate awareness. Dr. Queiroz worked on a similar project at Stanford’s Virtual Human Interaction Lab, focused on ocean acidification. Through explanations of cognitive psychology, the balance between interaction and narration, and importance of experience personalization, Dr. Quieroz helped us streamline our outlines for the steps, duration, and specifics of VR development and experiment processes. With Quieroz’ development pointers in mind, we turned to the platform Unity. We first researched other assets that people had created, to incorporate them into our own design. This design decision allows us to effectively use our time and resources to focus on creating and coding the environment rather than learning a brand new software for 3-D modeling. After selecting our assets, we compared the models of the molecules to stick models of the actual pollutant particles for a more accurate depiction. Subsequently, we drafted a static model with the city and pollutants. In the future, we plan on adding interactivity to the molecules so that users can learn about the pollutants in an engaging, and thus lasting, manner. In doing so, we aim to effectively minimize the disjunction between ourselves and our environment.
Metagenomic Analysis of a Compost Microbiome
 by Graham Houghton
by Graham Houghton
Metagenomics is the study of genetic material taken from populations of organisms. By performing metagenomic analysis of DNA from the Pingry composter, we aim to understand how the microbial community of the composter responds to changes in environmental conditions. In doing so, we hope to gain practical insights into the biological mechanisms which underlie the composting process. From the 2019-20 school year through the present, the team has successfully isolated and sequenced the DNA from 39 composter samples, yielding data on the relative abundances of
different types of bacteria within their microbial communities as shown in Figure 1. This year, we also used macromorphological analysis to investigate the presence of specific microbes in certain samples. Preliminary analysis of the data reveals differences between microbial communities present at different stages of the decomposition process. The analysis also reveals that several characteristics in the environment of the composter, such as temperature, pH, and amount of raw material added, have varying degrees of correlation to the abundance of certain bacterial orders.
Optimizing the Production of Artificial Collagen for use as a Biomaterial
 by Alexandra Drovetsky (VI), Ria Govil (IV), Sarina Lalin (V), Kirsten Lytle (VI), Annabelle Shil-
Theodore Strelecky (V) Advisors: Dr. P. Haven, Dr. M. D’Ausilio
by Alexandra Drovetsky (VI), Ria Govil (IV), Sarina Lalin (V), Kirsten Lytle (VI), Annabelle Shil-
Theodore Strelecky (V) Advisors: Dr. P. Haven, Dr. M. D’Ausilio
Collagen is the most abundant protein in the human body where it plays an important structural and functional role in many physiological and pathogenic processes. At the molecular level, collagen is made up of three polypeptide chains that form a secondary triple helical structure, which then self-assembles in a lateral staggered association to create fibrils with a unique 67 nm gap-overlap repeat known as D-period. The unique conformity and behavior of the collagen molecule arises from the repeating Glycine-X-Y pattern in its primary structure. The current goal of our project is to maximize the yield of collagen mimetic peptides that model the physical and chemical properties of natural type I collagen. We are currently utilizing a pre-existing peptide, named V-F877, which is 222 amino acids
long and has a V-domain, a trimerization domain found in bacteria that is known to optimize artificial collagen production. In order to better understand the role of the V-domain in improving the yield of collagen, we will compare the yield of the V-F877 sequence with and without the V-domain. Currently, collagen is most commonly purified from animals for use in the medical field. This process is expensive and often results in a high degree of variation. Generating collagen mimetic peptides that successfully replicate human collagen could provide a safer, pathogen-free, and cost-effective alternative. We plan to provide critical research on these foundational steps in order to deepen knowledge of collagen’s structure as well as determine the best methods of creating viable artificial collagen that can be used as a biomaterial.
PETase
by Juliana Amorosi (V), Maya Khan (VI), Sloane Mandelbaum (IV), Sebastian Talarek (V), Sofia Wood (IV) Advisors: Dr. M. D’Ausilio, Dr. J. Pousont
The United States only recycles about 35% of its total waste, sending nearly 150 million tons of trash to landfills each year. There, it takes up to 450 years to fully biodegrade. Often, it ends up invading oceans or forests, where it poses a serious threat to native species. Polyethylene terephthalate (PET) is one of the most widely used plastics in the United States, but only 31% of the 3.1 million tons of PET produced in America annually are recycled. It is necessary to find an efficient way to recycle plastic in order to mitigate the damage done to our planet. Our team works with PET depolymerase (PETase), an enzyme with the unique ability to break PET polymers into monomers, which can then be reassembled into new plastic. This nuanced recycling method is termed “biochemical recycling” and has become an increasing focus of research— however, it requires optimization before it can be implemented commercially. Because the severity and pervasiveness of plastic-related damage to the environment is growing, the scientific community is eagerly seeking to improve PETase through mutation–specifically, its efficiency and thermostability. Our goal is to identify and investigate various mutations, and assess the change in efficiency and thermostability that each mutation offers. After researching mutations that demonstrate promise for accomplishing these two goals through scientific literature, we obtained various PETase mutations through our partnership with Deerfield Academy. Deerfield created these mutations using site-directed mutagenesis, a common method to induce mutations, utilizing a primer to confer a mutation in double-stranded plasmid DNA. To replicate the mutated plasmid, we transform it into NEB 5-alpha cells and grow colonies on LB-AGAR plates with ampicillin. We harvest the colonies using inoculation loops before miniprepping: lysing the cells and discarding all contents besides the mutated
plasmid. Then, we transform the mutated DNA into BL21 cells and induce protein expression with autoinduction to create copies of the mutated PETase enzyme. Finally, we perform cell lysis and purification to obtain the pure enzyme. To determine the efficacy of mutant PETase,we plan to use a spectrophotometric assay or PCL-clearing assay. Either experiment will reveal whether, and how efficiently, our PETase mutants are capable of biodegrading PET plastic. By comparing the rate of degradation to our Wildtype control, we will find out which mutant is the most efficient. In the coming months, we hope to make more progress researching and documenting the effects of mutation on PETase activity, which will further understanding of the enzyme, and contribute to the global effort to make recycling more efficient and widely used.
Protein Purification of Shigella dysenteriae Transcription Factor VirF
 by Maddie Humphreys (VI), Daniela Karnaugh (V), Katia Krishtopa (V), Maria Loss (VI), Krish Patel (IV), Anika Sinha (V)
by Maddie Humphreys (VI), Daniela Karnaugh (V), Katia Krishtopa (V), Maria Loss (VI), Krish Patel (IV), Anika Sinha (V)
Advisors: Dr. J. Pousont, Dr. M. D’Ausilio
Abstract
Shigella is a pathogen that is a leading cause of bacterial foodborne illness worldwide. The virulence cascade of shigellosis requires one protein of the AraC superfamily transcription factors, VirF. Other members of this superfamily, such as ToxT, which is involved in the virulence cascade of Vibrio cholerae, contains a structural pocket where a small fatty acid attaches to inhibit DNA binding. We wish to determine whether or not VirF operates similarly, and thus be inhibited just as ToxT might. Therefore, we are working to express, purify, and ultimately solve the crystal structures of VirF and use biochemical property testing to assess for homologous function. Solving the structures will help to reveal the mechanism of transcriptional regulation in S. dysenteriae and could lead to development of novel treatments for Shigella related illnesses.
Introduction
The Shigella bacterium causes shigellosis, specifically mammalian gastroenteritis. An estimated 80-165 million cases of gastroenteritis due to Shigella species occur globally each year. Symptoms vary from inflammation of the intestines causing diarrhea to nausea and vomiting. Shigella also kills over 600,000 people annually. Infection begins with the ingestion of contaminated food or water. Many of the genes required for intestinal penetration, invasion of host cells, and intestinal and diarrheal disease are carried out by the major regulatory protein VirF located on the PINV as part of the regulatory cascade. The member of the AraC protein family is a DNA-binding protein that activates toxicity factors icsA and virB genes. These genes are then able to activate other downstream effectors involving invasion of the cell wall and host infection. Thus, inhibiting VirF binding to DNA would inhibit the whole virulence cascade that causes shigellosis. We wish to examine crystallized VirF and view the structural mechanism of DNA binding. We also wish to contrast VirF to ToxT to better understand the biochemical function.
Methods
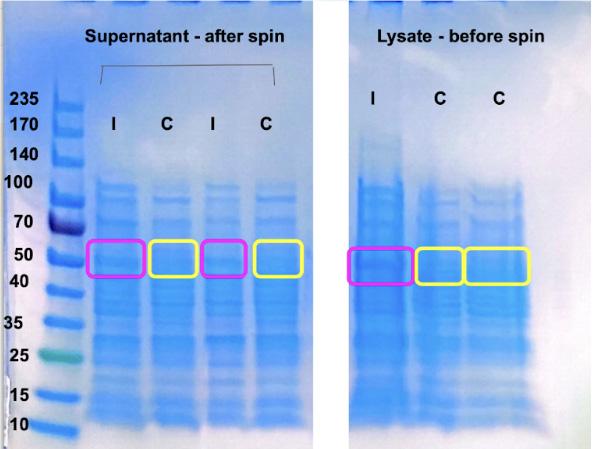
We received the VirF plasmid dried on filter paper from Dartmouth’s Kull Laboratory in January 2021. We rehydrated the plasmid in distilled water, and proceeded to conduct bacterial transformation to replicate the plasmid. Then, through a miniprep, we isolated
the plasmid from the bacteria and sent the plasmid to a lab for sequencing. We then analyzed the sequencing/plasmid map to confirm there are no mutations in our sample. After confirmation of correct plasmid we ran autoinduction to express protein and lysis to isolate VirF specifically from the cells (Figure 1).
Future Steps
Our goal is to isolate the VirF proteins using affinity chromatography purification and biochemical analysis to understand the mechanism of transcriptional regulation in S. dysenteriae. Understanding how to inhibit this protein could lead to development of novel treatments for Shigella related illnesses. Our next step is to perform batch purification to isolate our protein from bacterial cells using the intein tag attached to our plasmid. We will perform a gel to determine the success of our protein expression and purification. Once we have sufficient protein concentration, we intend to crystallize VirF and analyze the protein function via biochemical property testing. We hope to compare the structure of VirF to the previously solved structure of ToxT. Biochemical testing of VirF, including DNA binding assays and evaluation using EMSA analysis, will help to identify the function of VirF and its relationship to the homolog, ToxT.
ShallowMind
by Shaan Lehal (VI), JingJing Luo (IV), Alexander Recce (IV), Divya Subramanian (V), Olivia Taylor (VI), Evan Xie (V), Leo Xu (VI) Advisor: Dr. M. JollyAs artificial neural networks become increasingly ubiquitous in emerging technologies across a vast array of industries, the efficiency and accuracy of these machine learning tools are of critical importance. It is widely accepted that the structure of neural networks has an impact on their ability to interpret data, but the nuanced implications of network topology are largely not understood. The ShallowMind project seeks to answer this central question: how, and to what extent, does the structure of a neural net affect its efficiency and accuracy?
Previously, we have explored this question by building evolutionary networks and determining their efficacy compared to traditional neural networks. While traditional neural networks have a fixed structure and alter their weights and biases throughout training, evolutionary networks (ENN) undergo a rigorous process similar to that of biological evolution that not only optimizes weights and biases, but also network structure. The genetic algorithm dictating this optimization, “NeuroEvolution of Augmenting Topologies” (NEAT), reproduces, mutates, and trains a population of unique networks until a network performs sufficiently well on validation data. However, in many classification tasks, NEAT was unable to sufficiently evolve and achieve a higher efficacy than traditional neural networks.
This year, we changed the direction of our research to focus on NEAT’s most common real-world application, video game AI, in hopes of demonstrating improved efficacy over traditional neural networks. First, we tackled the Google Dinosaur Game, which progressively becomes more
difficult as the player survives and earns a higher fitness. Most recently, we have been applying NEAT to the Game of Pure Strategy in which two players try to outbid each other with card values.
So far, the implementation of NEAT to the Dinosaur game has yielded three main findings. First, when NEAT evolves to find the solution to a game, it typically evolves randomly instead of linearly. Second, when NEAT fails to solve a game, it instead finds and sticks to a local solution. For the Google Dinosaur Game, this meant that the dinosaur either never jumped or always jumped. Third, the NEAT hyperparameters pertaining to speciation and structural mutation, which dictate NEAT’s evolution, do not have a statistical significance on NEAT’s performance.
Through the Game of Pure Strategy, we have demonstrated that NEAT can learn the optimal strategy for a game with a “human element” against both an opponent playing randomly and an opponent with a clear strategy. In the future, we hope to train a network which can determine an opponent’s strategy during the game and adapt to it as the game goes on, rather than needing to be trained against a specific behavior in order to excel against it.
In the future, we hope to apply NEAT to other games similar to the Game of Pure Strategy which require complex strategic behavior and a “human element,” which can’t be won simply with memorization.
Smart Heart: Deep Learning-Based Analysis of 12-Lead Electrocardiograms for Cardiovascular Disease Risk Prediction
by Elbert Ho (IV), Max Liu (VI), JP Salvatore (VI), Vinav Shah (IV), Evan Wen (VI), Alan Zhong (V)Advisor: Dr. M. Jolly
Cardiovascular disease is the leading cause of death worldwide and is often diagnosed with electrocardiograms (ECG). The electrocardiogram is a non-invasive test that measures the heart’s electrical signal via electrodes placed on the chest and limbs. Diagnosing ECGs was originally a task left to medical specialists, but in recent years, AI has emerged as an approach to automate the diagnosis. However, there are limitations leading to various inaccuracies that, to this date, AI has been unable to conquer fully. Our project is focused on solidifying this approach with optimal diagnosis results. We explore various signal processing methods to denoise our data, such as finite impulse response, discrete wavelet transforms, feature engineering, and Fourier transform.
While reading other teams’ code and literature from the 2020 George B. Moody PhysioNet Challenge, we found it difficult to add new features to improve their results. This year, we created a framework encompassing all aspects of the ECG diagnosis process, from data loading and preprocessing to model training and evaluation. Our
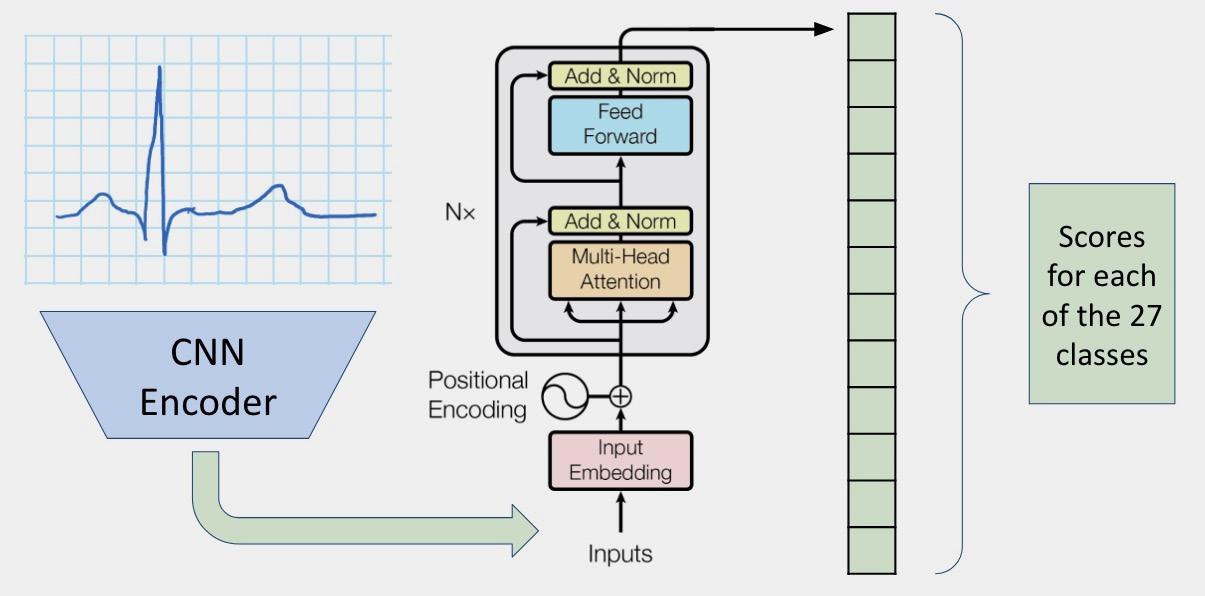
pipeline makes it easy to swap in interchangeable parts, allowing us to add more features to our code. We used two different approaches to develop our model, stemming from the two most popular fields in deep learning: natural language processing and computer vision. Inspired by works in natural language processing, we developed an architecture that incorporates features learned by a convolutional neural network. Then, these features are fed into popular natural language processing architectures such as LSTM, GRU, and transformers. From the field of computer vision, we adapted deep convolutional neural network architectures that we designed to perform well on visual tasks such as image classification, image segmentation, and object recognition to denoise and extract features from ECG data.
Our results are evaluated against other approaches from the 2020 PhysioNet Challenge, which featured 41 competing teams. The performance of our models is comparable to the top 10 teams from the official contest.
Studying Zooxanthellae to Protect Coral
 by Emily Gao (V), John Grissinger (VI), Matthew Oatman (VI), Sejal Patel (VI), Saniya Tariq (V), Melinda Xu (IV), Rohan Yadav (IV)
by Emily Gao (V), John Grissinger (VI), Matthew Oatman (VI), Sejal Patel (VI), Saniya Tariq (V), Melinda Xu (IV), Rohan Yadav (IV)
Advisor: Ms. S. Mygas
Coral polyps have a symbiotic relationship with zooxanthellae, which are photosynthetic dinoflagellates that help corals absorb nutrients from their environment. Zooxanthellae are responsible for providing the coral with the requisite nutrients for survival, and typically assist the process of photosynthesis (NOAA).

Zooxanthellae play a major role in protecting and maintaining coral reefs; however, when there is a depleted amount of zooxanthellae present in the coral reefs, the coral begins to bleach and stress (NOAA). Our research team aims to extract and understand zooxanthellae, so we can design a solution that will prevent premature coral reef death and increase coral reef longevity. If we can uncover patterns and trends in the change of zooxanthellae during horizontal transfer, among individual corals housed near each other, we can make inferences about how global warming is affecting coral reefs in our oceans, and we plan to share our discoveries among the community. In order to uncover these trends, we will take samples of the coral and extract the zooxanthellae. We will then fingerprint their DNA in order to
record the various species of zooxanthellae present in the coral. Once we establish a baseline library of zooxanthellae identities and quantities, we will develop further questions and parameters for measuring zooxanthellae community dynamics.
Thus far, the coral group has designed a process for tank maintenance. We first measured a tank size and depth that would be habitable for our coral, and after obtaining an adequate tank, our group began developing a procedure to form a habitable system. Our weekly procedures incorporate a mechanism for tank maintenance. We start by checking the pH level to confirm the accurate acidity of the water. We then check the salinity to ensure that there is sufficient salt in the water — our desired salinity is around 35 ppt. Based on the salinity of the tank water, we then change the water with a newly created saltwater substance. Our group has now received our coral and we have formulated a feeding technique that would be safe for the coral prior to commencing our planned research.
SMART Team.
The Dnase1L3 Flexible C-Terminal Domain and Altered Charges in the Actin Recognition Site Result in Broad Substrate Diversity and Resistance to Actin Inhibition
by Sasha Bauhs (IV), Olivia Buvanova (IV), Ben Chung (IV), Mia Cuiffo (IV), Jiya Desai (IV), Kayla Kerr (IV), Valentyn Kurylko (IV), Sienna Patel (IV) Advisor: Dr. J. PousontIn the United States alone, 1 in 15 people are affected by autoimmune diseases. These illnesses are caused when body tissue is attacked by its own immune system, which can occur due to the build up of extracellular DNA after cell death. Most extracellular DNA is cleared by nonspecific endonucleases Dnase1L3 and Dnase1. Dnase1L3 activity, when compromised, cannot be rescued by the presence of Dnase1. While the cellular activity of Dnase1 is heavily inhibited by actin, Dnase1L3 is more proficient in degrading extracellular DNA because it resists actin inhibition. Additionally, Dnase1L3 degrades antigenic forms of DNA, including protein-DNA and lipid-DNA complexes, that Dnase1 cannot degrade. Decreased function of Dnase1L3, even with fully functioning Dnase1, can lead to the development of autoimmune diseases. For instance, the R206C mutation reduces activity of Dnase1L3 and is linked to the development of lupus.

To examine the structural basis of the enhanced activity of Dnase1L3 in comparison to Dnase1, researchers crystallized the core domain of Dnase1L3 (Dnase1L3 ΔCTD). The last 23 residues, comprising the flexible C-terminus (CTD), were removed to facilitate crystallization. Several key structural differences are responsible for its resistance to actin. In the central hydrophobic region of Dnase1L3, a bulky Phe-131 disrupts actin binding. An out-of-register ��-helix and the insertion of amino acids in a loop following this helix result in loss of salt bridge formation and electrostatic repulsion of actin, respectively. The CTD of Dnase1L3 enhances degradation of DNA that is complexed with lipids or antibodies. Researchers examined the structure of the
CTD using Nuclear Magnetic Resonance spectroscopy and then modeled this domain together with the core domain using Small-Angle X-ray Scattering. The CTD was found to be a flexible, disordered DNA-binding domain that projects outward from the core domain. It was proposed that the CTD enhances Dnase1L3 degradation of diverse forms of DNA because of its structural flexibility.
The Pingry School SMART team used the 3-D modeling program, Jmol, to design a model of Dnase1L3 in order to better understand how the unique structure of Dnase1L3 correlates to its function. This model was custom printed by 3D Molecular Designs. Important differences compared to Dnase1 have been highlighted in our model, and allow for a clearer visualization of the structural features of Dnase1L3 that lend it its enhanced DNA degradation functions. A fuller understanding of the unique structure of Dnase1L3, including the CTD and actin recognition site, will allow Dnase1L3 based therapies to be created to treat autoimmune diseases.
Club Spotlights.
Robotics Club
Dwayne Bazil (VI), Jesse Busch (III), Keira Chen (V), Matt Dicks (IV), Rayee Feng (III), Sia Ghatak (IV), James Kotsen (IV), Laura Liu (V), Diego Pasini (VI), Aadi Stewart (III), Olivia Taylor (VI), Alan Zhong (V), Carolyn Zhou (IV), Christian Zhou-Zheng (III)
byDo you like robots? Of course you do! Come see Pingry Robotics in action by interacting with our world-championship robots; they can do pull-ups, climb stairs, make half-court shots, raise discs ten feet in the air, and more! At our research week exhibit you will be able to drive some of our robots, see our robots that got to world championships, and ask questions to the brilliant team behind them.
The Pingry robotics team builds two robots to complete specific tasks in time-constrained competitions to win points. We compete in FTC (FIRST Tech Challenge) and FRC (FIRST Robotics Competition) along with mentoring the middle school FTC team.
In this year’s FTC competition, points are scored by placing cones onto flexible yellow rods of varying heights. Our robot this year has a horizontally extendable claw, which is attached to a set of vertically extending slides. The slides are attached to a rotatable turret, allowing us to reach lots of different heights and locations without moving the chassis of the robot. Many parts of our robot, specifically much of the vertical slides and claw, were custom designed and 3-D printed. We experimented with different materials, ultimately settling on PETG for strength to better protect our robot from breaking during competition. During the autonomous period of the match (the first 30 seconds), the robot drives itself using sensors and cameras which help it move and score, and during the tele-operated period (the remaining 2 minutes), our drivers use gamepads to control the robot. While autonomous, our robot uses computer vision to determine which of three QR codes on a sleeve of a cone are facing it, and then drives to different places depending on the cone. In this year’s FRC competition, points are scored by placing cones and inflatable cubes onto three different levels, that
each have fixed rods on them. In addition, points are awarded for being able to balance multiple robots on a moving platform. Our robot has a rotating extending arm and a special drivetrain that allows for faster and more accurate maneuverability. Additionally, we have pneumatics that power our claw.
Throughout the past few years we have been using our skills and experience from FTC and FRC in order to help those around us. We have embraced Pingry’s values of intellectual engagement, discovery, and innovation to help us continue building amazing robots.
Anatomy Club
by Lauren Kim (VI), Sarina Lalin (V), Mia Libretti (V), Maria Loss (VI)Anatomy Club meets every other month to dissect various specimens, including animals and organs. So far, we have dissected fetal pigs, sharks, snakes, hearts, brains, and more. Dissections are a fun and hands-on way to learn about anatomy, especially for those interested in pursuing a career in the biomedical field. Through this club, we hope to provide memorable learning experiences that instill in students a love for science and exploration.
FYI Sci
by Maria Loss (VI), Sophia Odunsi (V)Formerly known as Project 80, FYI Sci is a student-led science organization. Our goal is to share science and its wonders with the Pingry community. FYI Sci shows everyone how fun science can be through blogs, videos, and community events. In the past, FYI Sci has primarily been a thrill during Pingry’s research week, with activities every day throughout campus. During that week, FYI Sci always gets the community involved in egg drop competitions, trivia games, and strawberry DNA extractions to support their mission. Be on the lookout for this year’s fun and science-filled research week, and please check out our website (https://students.pingry.org/fyisci/) to stay up to date with our resources.

Email:
Evan Xie (V): exie2024@pingry.org



Annabelle Shilling (V): ashilling2024@pingry.org
Mr. D. Maxwell: dmaxwell@pingry.org




