
15 minute read
Ishaan Goswami
A Bioinformatic Analysis of mecA Prevalence in Bacterial Species
BY ISHAAN GOSWAMI (BAYVIEW GLEN INDEPENDENT SCHOOL)
Advertisement
Cover Image: View of double stranded DNA
Image Source: Pixabay Abstract: Antibiotic resistance is a serious issue in healthcare. mecA is the gene in MRSA and many other bacterial species responsible for the development of resistance to commonly used β-lactam antibiotics. MRSA and other β-lactam resistant bacteria pose risks to health by increasing healthcare related mortality and morbidity rates. This paper aims to display the promising usage of bioinformatic tools in tackling antibiotic resistance by investigating the prevalence of mecA across bacterial species. It is imperative that further research be done to investigate the prevalence of other antibiotic resistant genes using similar bioinformatic approaches.
Introduction Antibiotic resistance is a problem of great concern for the medical community. “Superbugs” are pathogens that undergo mutations which causes resistance to treatments, and can cause increasingly harmful infections. One such superbug is Methicillin-resistant Staphylococcus aureus (MRSA). Staphylococcus aureus is a gram-positive, coccal bacteria which causes staph infections and has the potential to cause serious complications, such as pulmonary, coronary, and urinary tract infections (Taylor & Unakal, 2018). Due to mutations, a strain of S. aureus has developed into MRSA and has become resistant to commonly used β-lactam antibiotics, which include penicillin and its derivatives such as methicillin (Green et al., 2012).
mecA is a gene in MRSA conferring resistance to β-lactam antibiotics. (Ubukata et al., 1989). In susceptible strains of S. aureus, β-lactam antibiotics specifically inhibit a transpeptidase that catalyzes cell-wall crosslinking between polysaccharide chains and peptides, which creates a strong polymer called peptidoglycan that maintains the rigidity and shape of bacterial cell walls. However, in mutated forms of S. aureus that contain mecA, the gene encodes for a protein called PBP2A (penicillin-binding protein 2A), which has a low affinity for β-lactam antibiotics. This means that it does not bind to β-lactam antibiotics. This means that cell wall synthesis is not inhibited, and bacterial reproduction is unhampered (Fishovitz et al., 2014).
MRSA is not benign and has been observed in clinical settings to be hazardous to health. Typically, MRSA infections occur in hospitals, nursing homes, dialysis centers, and other primary care locations. These cases of MRSA are called healthcare associated MRSA, or
HA-MRSA. MRSA can be contracted during surgeries, or by touching unclean surfaces (Mayo Clinic, 2018). Community-associated MRSA, or CA-MRSA can spread by skin-to-skin contact between seemingly healthy populations. Thus, people who are constantly in physical contact with other people such as wrestlers, or people living in crowded conditions are considered at risk populations (Mayo Clinic, 2018).
The main unsolved issue surrounding MRSA is the lack of effective remedies to infections. Since methicillin is a narrow range antibiotic, meaning it can only inhibit a limited number of bacterial species, it has become increasingly difficult to treat such infections. Since methicillin targets specific bacteria, antibiotics that would have targeted these infections become unresponsive. Thus, broad range antibiotics must be employed, which is discouraged as they can develop antibiotic resistance in more bacterial species. MRSA is responsible for approximately 80,461 infections, and 11,285 deaths annually in the United States (Melander et al., 2017). It is evident that MRSA poses a serious threat to public health.
Two bioinformatic analyses related to the mecA gene from MRSA were conducted in this paper; these included the investigation of the spread of mecA to other bacterial strains via homologybased searches and exploration available information about the structure and function of mecA protein via secondary databases. This work will hopefully improve the understanding of the characteristics and spread of mecA and will thus contribute to the efforts of combating MRSA.
Methods Investigating the prevalence of mecA across bacterial species
To investigate the prevalence of mecA, and analogous proteins across bacterial species, BLAST was utilized (using the accession number “MW682923”), which was accessed at the National Centre for Biotechnology Information (NCBI) website. BLAST stands for basic local alignment search, and is an algorithm used to compare primary biological sequence information. BLAST compares a subject sequence called a query against a database of sequences to find analogous matches. BLASTn was used for the nucleotide search, while BLASTp was used for the protein search.
For hits to be considered biologically significant, the following cut-offs, based on current literature (Lee et al., 2010; Kaur et al., 2019), have been introduced to the BLAST searches: >50% Percentage Identity, >80% coverage, and E-value <1e-5.
Investigating the function of mecA
To investigate the function of mecA, public secondary databases were employed. uniProt, a secondary database which contains protein annotations; Pfam, a secondary database which analyzes protein sequences and determines conserved protein domains; and Phobius, a secondary database that analyzes protein signatures to figure out where in the cell they are located. "The main unsolved issue surrounding MRSA is the lack of effective remedies to infections."
Image 1: Bioinformatic tools can effectively be used to track antibiotic resistance
Image Source: Flickr
Results
Investigating the prevalence of mecA across bacterial species
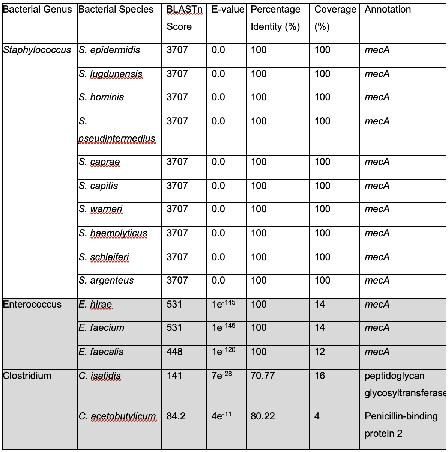
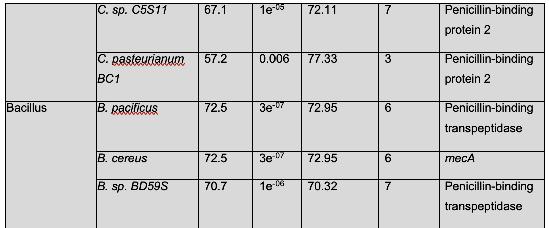
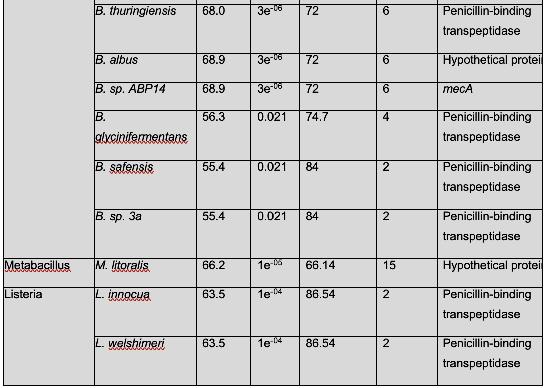
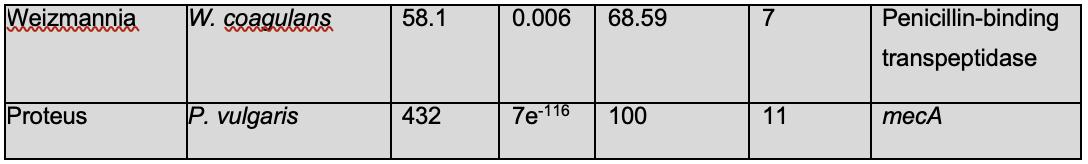
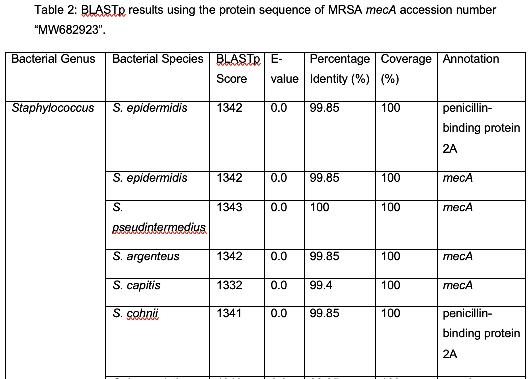
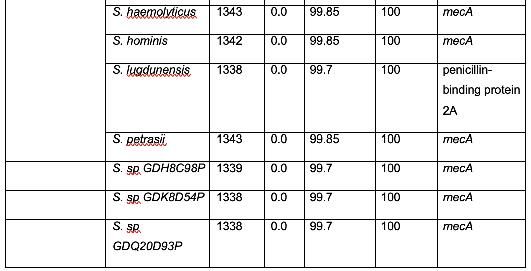
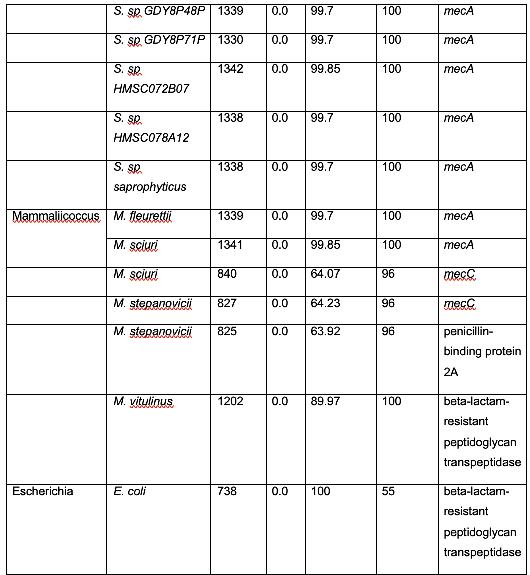
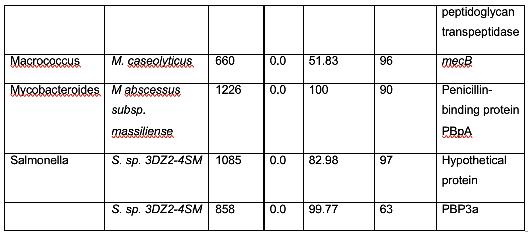
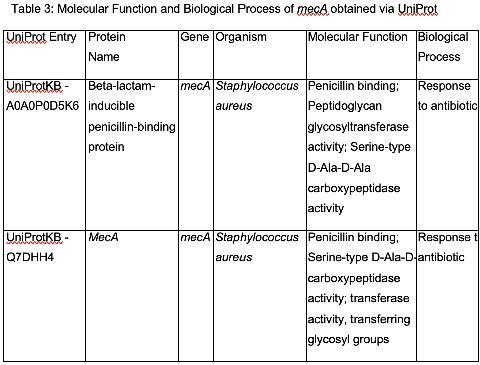
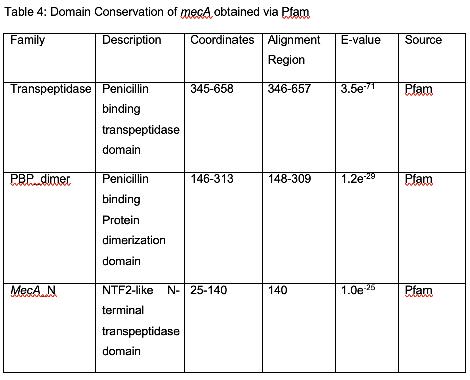
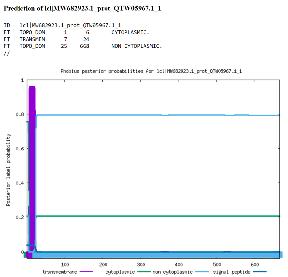
Image 2: Phobius Posterior Proabilities for mecA Source: Produced by Goswami via Phobius
Discussion Investigating the prevalence of mecA across bacterial species
Table 1 shows that only bacteria of the genus Staphylococcus met the required cut-offs to be considered significant. This means that all the species that were hits in the BLASTn search have mecA, and these findings are biologically significant. This is in line with previous labbased experimentation, which finds that mecA exists in multiple other Staphylococcal species found in humans and animals called coagulasenegative staphylococci (CoNS) (Becker et al., 2014). BLASTn only accounts for hits with regards to the genetic sequence of the query and other organisms, not the function or expression of this sequence. However, even though some of the species do not meet the cut off, they should be considered and investigated, as on the nucleotide level, they may be different, while they create proteins with similar structures and functions. The hits obtained that did not meet the cut-offs had annotations of peptidoglycan glycosyltransferase, mecA, penicillin-binding protein 2, and penicillin binding transpeptidase, all of which are either mecA or use the similar mechanisms to obtain β-lactam resistance.
An analysis of the results of the BLASTn hits shows multiple bacterial species that have β-lactam resistance. Enterococci have been shown to have β-lactam resistance by altering penicillinbinding proteins (mecA) and the production of a β-lactamase in E. faecalis (Fontana et al., 1990). Genus Clostridium has been shown to have developed β-lactam resistance by producing β-lactamase like E. faecalis (Sandhu et al., 2019). Genus Listeria has been observed to have an innate resistance to β-lactam antibiotics (Krawczyk-Balska et al., 2012). The two hits both employ Penicillin-binding transpeptidase, which is thus an intrinsic part of its genome, and not a mutation. P. vulgaris has been shown to exhibit β-lactam resistance.
There are certain novel findings that must be examined in further detail. M. litoralis has not been experimentally confirmed to possess β-lactam resistance, but these findings show that it should be investigated further via experimentation. W. coagulans has been shown to be a probiotic agent in dairy, but there is no literature on this species’ β-lactam antibiotic resistance (Tóth et al., 2021). Dairy is a consumer product, thus, W. coagulans has the potential to be a public health concern. It is implored that this finding is observed in more detail.
Table 2 shows organisms that have a similar protein sequence to the query. Staphylococcus, Mammaliicoccus, Escherichia, Listeria, Macrococcus, Mycobacteroides, and Salmonella are the represented genera in this BLASTp. However, it should be noted that most of these are not annotated as mecA. Instead, they are typically a penicillin binding protein, beta-lactam resistant peptidoglycan transpeptidase, mecB, or mecC. This shows that these are analogous proteins, but not identical to mecA. Their amino acid sequence is similar, and thus, in these cases, have similar functions. It has been shown that non-staphylococcal bacteria, such as E. coli have gained β-lactam antibiotic resistance, which corroborates the results found in this analysis. (Kassem et al., 2008)
E. coli has been experimentally observed to have gained antibiotic resistance to β-lactam antibiotics and has shown the potential to develop further antibiotic resistance via laboratory cloning (Jacoby & Sutton, 1985). Listeria, as stated previously, has been discovered to have
innate β-lactam resistance (Krawczyk-Balska et al., 2012). Salmonella has also been associated with β-lactamase production, which provides β-lactam resistance (Souza et al., 2020).
There were also several findings of note. Genus Mammaliicoccus has been shown computationally to have β-lactam resistance. However, the only information available on this genus are primary database entries, and a study on wide-range antibiotic resistance in various Mammaliicoccus species in cattle (Schauer et al., 2021). This result must be investigated further. M. caseolyticus have been shown to carry mecB, as these results show, and mecD, which is a novel gene that shares a 66% nucleotide identity with mecB (Schwendener et al., 2017). This novel gene is a potential area of further inquiry into the evolution of antibiotic resistance. M. abscessus has innate antibiotic resistance. However, it has been determined that a combination of β-lactam antibiotics have a 100% efficacy rate against M. abscessus (Story-Roller et al., 2021). This is encouraging, as M. abscessus is associated with Penicillin-binding protein PBpA, a β-lactam resistance conferring protein.
These results show that members of the genus Staphylococcus other than MRSA are genetically homologous for mecA, while other bacterial genera exhibit β-lactam antibiotic resistance, which are analogous in terms of protein sequence to mecA.
Investigating the function of mecA
Table 3 shows the protein annotations of mecA. Penicillin binding is in line with the function of mecA and PBP2A. D-Ala-D-Ala carboxypeptidase activity entails the activity of an enzyme that is involved in cell wall integrity and strength (Spidlova et al., 2018). Β-lactam antibiotics work by inhibiting cell wall growth, and thus reproduction. The D-Ala-D-Ala carboxypeptidase activity thus counteracts this. Peptidoglycan glycosyltransferase activity involved in biosynthesis of bacterial cell wall, involves enzyme reactions that take place within the cytoplasm, and outside the cytoplasmic membrane. (Barreteau et al., 2008). This is in line with the methodologies of PBP2A to counteract Β-lactam antibiotics. The transmembrane nature of Peptidoglycan glycosyltransferase activity is in line with results from the Phobius analysis.
Table 4 shows the conserved domains of mecA. Domains are more conserved evolutionarily than other regions of a protein and evolve as units. Domains are also structurally distinct from the rest of the protein and have their own functions. These domains can be used to infer function, and proteins which probably have similar functions, but this must be tested via experimentation. There are three conserved domains: Penicillin binding transpeptidase domain; Penicillin binding Protein dimerization domain; and NTF2-like N-terminal transpeptidase domain.
Transpeptidase is responsible for catalyzing cell wall cross-linking, which ensures bacterial cell wall integrity (Fishovitz et al., 2014). Thus, the Penicillin binding transpeptidase domain is related to maintaining cell wall structure and integrity. Dimerization is a process wherein two chemically similar molecules form a dimer, which is a single polymer. Dimerization is important role in maintaining transmembrane proteins. Dimerization is important for polymerizing peptidoglycan, which is related to the function of PBP2A. Its transmembrane nature is also in line with the Phobius findings, which explains the Penicillin binding Protein dimerization domain. The NTF2-like N-terminal transpeptidase domain’s length suggests a possible structural role of the domain, and thus gives the transpeptidase domain a longer reach from the cell membrane (Lim & Strynadka, 2002). This means that it is involved in bacterial cell wall cross-linking and is thus a part of the function of mecA.
Figure 2 shows the probabilities of the location of mecA in a cell by analyzing protein signatures. The Phobius Posterior Probabilities for mecA clearly shows a transmembrane region (purple, score ≈ 1) and a non-cytoplasmic region (light blue, score = 0.8). These results show that it resides on the outside of the cell with a transmembrane region across the bacterial membrane, which is fully in line with the mechanism of PBP2A against β-lactam antibiotics, and with the findings from UniProt and Pfam.
An analysis of secondary databases may shine light on the structure, location, and function of mecA, and all these results are in line with the mechanisms of antibiotic resistance employed by PBP2A. A firm understanding of these functions and structures can be used to determine effective counters to these new antibiotic resistant species.
Conclusion
This bioinformatical analysis highlighted the use of bioinformatic tools to track antibiotic resistance in bacterial populations, and thus formulate adequate responses to this urgent issue. The novel findings of β-lactam antibiotic resistance in the species M. litoralis, W. coagulans and genus Mammaliicoccus must be investigated further. They represent a potential concern in healthcare, as new species of antibiotic resistant bacteria. More research into the analysis of analogous protein structures to ¬mecA in various bacterial species will be an important step to combatting antibiotic resistance from a bioinformatics perspective.
Acknowledgements
I would like to acknowledge Dr. Ashraf Zarkan of the University of Cambridge for his guidance and mentorship in this project. I would like to thank the Cambridge Centre for International Research for their sustained support throughout the research process. I would also like to thank Ms. Kristin Carpenter of the Bayview Glen Independent School for supporting my passion for, and exploration in the biological sciences.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of molecular biology, 215(3), 403–410. https://doi.org/10.1016/S00222836(05)80360-2 Barreteau, H., Kovač, A., Boniface, A., Sova, M., Gobec, S., & Blanot, D. (2008). Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiology Reviews, 32(2), 168–207. https:// doi.org/10.1111/j.1574-6976.2008.00104.x Becker, K., Heilmann, C., & Peters, G. (2014). Coagulase-Negative Staphylococci. Clinical Microbiology Reviews, 27(4), 870–926. https:// doi.org/10.1128/cmr.00109-13 EBI. (n.d.) Sequence: MW682923.1. Www.ebi. ac.uk. https://www.ebi.ac.uk/ena/browser/view/ MW682923 Fishovitz, J., Hermoso, J. A., Chang, M., & Mobashery, S. (2014). Penicillin-binding protein 2a of methicillin-resistantStaphylococcus aureus. IUBMB Life, 66(8), 572–577. https://doi. org/10.1002/iub.1289 Fontana, R., Canepari, P., Lleò, M. M., & Satta, G. (1990). Mechanisms of resistance of enterococci to beta-lactam antibiotics. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology, 9(2), 103–105. https://doi. org/10.1007/BF01963633 Green, B. N., Johnson, C. D., Egan, J. T., Rosenthal, M., Griffith, E. A., & Evans, M. W. (2012). Methicillin-resistant Staphylococcus aureus: an overview for manual therapists. Journal of Chiropractic Medicine, 11(1), 64–76. https://doi.org/10.1016/j.jcm.2011.12.001 Jacoby, G. A., & Sutton, L. (1985). Beta-Lactamases and beta-lactam resistance in Escherichia coli. Antimicrobial Agents and Chemotherapy, 28(5), 703–705. https://www.ncbi.nlm.nih.gov/pmc/ articles/PMC176364/#:~:text=Escherichia%20 coli%20strains%20determining%2017 Käll, L., Krogh, A., & Sonnhammer, E. L. L. (2004). A Combined Transmembrane Topology and Signal Peptide Prediction Method. Journal of Molecular Biology, 338(5), 1027–1036. https:// doi.org/10.1016/j.jmb.2004.03.016 Kassem, I. I., Esseili, M. A., & Sigler, V. (2008). Occurrence of mecA in Nonstaphylococcal Pathogens in Surface Waters. Journal of Clinical Microbiology, 46(11), 3868–3869. https://doi. org/10.1128/jcm.01035-08 Kaur, H., Bose, C., & Mande, S. S. (2019). Tryptophan Metabolism by Gut Microbiome and Gut-Brain-Axis: An in silico Analysis. Frontiers in Neuroscience, 13. https://doi.org/10.3389/ fnins.2019.01365 Krawczyk-Balska, A., Marchlewicz, J., Dudek, D., Wasiak, K., & Samluk, A. (2012). Identification of a ferritin-like protein of Listeria monocytogenes as a mediator of β-lactam tolerance and innate resistance to cephalosporins. BMC Microbiology, 12, 278. https://doi.org/10.1186/1471-2180-12278 Lee, J.-H., & Lee, J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiology Reviews, 34(4), 426–444. https:// doi.org/10.1111/j.1574-6976.2009.00204.x Lim, D., & Strynadka, N. C. J. (2002). Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nature Structural Biology, 9(11). https://doi. org/10.1038/nsb858 Mayo Clinic. (2018). MRSA infection - Symptoms and causes. Mayo Clinic; https://www.mayoclinic. org/diseases-conditions/mrsa/symptoms-causes/ syc-20375336 Melander, R. J., Zurawski, D. V., & Melander, C. (2017). Narrow-spectrum antibacterial agents. MedChemComm, 9(1), 12–21. https://doi. org/10.1039/c7md00528h Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G., Sonnhammer, E. L. L., Tosatto, S. C. E., Paladin, L., Raj, S., Richardson, L. J., Finn, R. D., & Bateman, A. (2020). Pfam: The protein families database in 2021. Nucleic Acids Research, 49(D1), D412–D419. https://doi. org/10.1093/nar/gkaa913 MW682923. (n.d.). Getentry.ddbj.nig.ac.jp. http://getentry.ddbj.nig.ac.jp/getentry/na/ MW682923?filetype=html Sandhu, B. K., Edwards, A. N., Anderson, S. E., Woods, E. C., & McBride, S. M. (2019). Regulation and Anaerobic Function of the Clostridioides difficile β-Lactamase. Antimicrobial Agents and
Chemotherapy, 64(1). https://doi.org/10.1128/ AAC.01496-19 Schauer, B., Szostak, M. P., Ehricht, R., Monecke, S., Feßler, A. T., Schwarz, S., Spergser, J., Krametter-Frötscher, R., & Loncaric, I. (2021). Diversity of methicillin-resistant coagulasenegative Staphylococcus spp. and methicillinresistant Mammaliicoccus spp. isolated from ruminants and New World camelids. Veterinary Microbiology, 254(109005). https://doi. org/10.1016/j.vetmic.2021.109005 Schwendener, S., Cotting, K., & Perreten, V. (2017). Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Scientific Reports, 7(1). https://doi.org/10.1038/srep43797 Souza, A. I. S., Saraiva, M. M. S., Casas, M. R. T., Oliveira, G. M., Cardozo, M. V., Benevides, V. P., Barbosa, F. O., Freitas Neto, O. C., Almeida, A. M., & Berchieri, A. (2020). High occurrence of β-lactamase-producing Salmonella Heidelberg from poultry origin. PLOS ONE, 15(3). https:// doi.org/10.1371/journal.pone.0230676 Spidlova, P., Stojkova, P., Dankova, V., Senitkova, I., Santic, M., Pinkas, D., Philimonenko, V., & Stulik, J. (2018). Francisella tularensis D-Ala D-Ala Carboxypeptidase DacD Is Involved in Intracellular Replication and It Is Necessary for Bacterial Cell Wall Integrity. Frontiers in cellular and infection microbiology, 8(111). https://doi. org/10.3389/fcimb.2018.00111 Staphylococcus aureus strain MRSA-12 penicillinbinding protein 2 (mecA) gene, complete cds. (2021). NCBI Nucleotide. https://www.ncbi.nlm. nih.gov/nuccore/MW682923.1?report=fasta Story-Roller, E., Galanis, C., & Lamichhane, G. (2021). β-Lactam Combinations That Exhibit Synergy against Mycobacteroides abscessus Clinical Isolates. Antimicrobial Agents and Chemotherapy, 65(4). https://doi.org/10.1128/ AAC.02545-20 Staphylococcus aureus strain MRSA-12 penicillinbinding protein 2 (mecA) gene, complete cds. (2021). NCBI Nucleotide. https://www.ncbi.nlm. nih.gov/nuccore/MW682923.1?report=fasta Taylor, T. A., & Unakal, C. G. (2018). Staphylococcus Aureus. Nih.gov. https://www. ncbi.nlm.nih.gov/books/NBK441868/ Tóth, A. G., Csabai, I., Judge, M. F., Maróti, G., Becsei, Á., Spisák, S., & Solymosi, N. (2021). Mobile antimicrobial resistance genes in probiotics. BioRxiv. https://doi. org/10.1101/2021.05.04.442546 Ubukata, K., Nonoguchi, R., Matsuhashi, M., & Konno, M. (1989). Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureusspecific penicillin-binding protein. Journal of Bacteriology, 171(5), 2882–2885. https://www. ncbi.nlm.nih.gov/pmc/articles/PMC209980/ UniProt. (n.d.). mecA - Beta-lactam-inducible penicillin-binding protein - Staphylococcus aureus - mecA gene & protein. (n.d.). Www. uniprot.org. https://www.uniprot.org/uniprot/ A0A0P0D5K6 UniProt. (n.d.). mecA - MecA - Staphylococcus aureus - mecA gene & protein. (n.d.). Www. uniprot.org. https://www.uniprot.org/uniprot/ Q7DHH4










