
International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net p-ISSN:2395-0072


International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net p-ISSN:2395-0072
Almas1 , Chandana Y2, Iram Zahra3, Laiba Kounain⁴ , Anil Kumar C5
¹ ² ³ ´ UG Students, Department of Electronics and Communication Engineering, PESITM, Shivamogga, Karnataka, India
µ Assistant Professor, Department of Electronics and Communication Engineering, PESITM, Shivamogga, Karnataka, India ***
Abstract - Genomic data analysis deciphers the genetic code within DNA, providing insights into biology, diseases, and evolutionary patterns. By leveraging advanced sequencingtechnologiesandcomputationaltechniqueslike machine learning, researchers can identify genetic variations, analyze gene expressions, and conduct population studies. This field enables breakthroughs in personalized medicine, evolutionary studies, and agricultural improvements. We discuss methodologies such asvariantcalling,geneexpressionanalysis,andmulti-omics integration to extract meaningful insights from genomic data. These methods are revolutionizing healthcare, agriculture,andforensics.
Key Words: Genomic data analysis; Machine Learning; Bioinformatics; Biomarkers; Variant Analysis; Disease Research;GeneticStudy
Genomic data analysis plays a pivotal role in understanding the intricate biological processes encoded withinDNA.Asresearchersdelveintothegeneticcodesof organisms, they uncover valuable insights into genetic predispositions,diseasemechanisms,evolutionarytrends, and much more. The advancement of sequencing technologies, particularly Next-Generation Sequencing (NGS), has revolutionized this field, enabling the efficient sequencing of entire genomes, transcriptomes, and epigenomes. These innovations have resulted in massive datasets, necessitating the use of computational methods toprocess,analyze,andinterpretthedataeffectively.
The integration ofmachine learning (ML)techniques has emerged as a powerful tool for extracting meaningful patterns and making predictions from these complex datasets. Machine learning methods, such as supervised learning, unsupervised learning, and deep learning, allow researchers to identify genetic variations, classify species, and predict disease risks, providing insights that were previouslyunattainable.Fromgeneexpressionanalysisto the detection of genetic biomarkers for diseases, machine learningmodelshelpunravelthevastpotentialofgenomic data.
Inthispaper,weexplorethedifferentmethodologiesused in genomic data analysis, focusing on the role of machine learning in tasks such as variant calling, gene expression analysis,andmulti-omics integration. Theultimategoal is todemonstratehowthesecomputationalapproacheshave revolutionized fields like personalized medicine, agriculture, and evolutionary biology. We also discuss the challenges associated with large-scale genomic data, including data preprocessing, quality control, and ethical considerations, especially in medical applications. Through these discussions, we highlight how genomics, when combined with machine learning, is set to drive the next wave of scientific breakthroughs and innovations in varioussectors
Development ofa machinelearning-basedframework for DNA sequence analysis to address three major challengesingenomics:identificationofspecies,detection ofpromoterregions,andclassificationofDNAsequences.
1. DNA Sequence Classification: Develop machine learning modelsto classifyDNAsequencesinto seven predefined functional or structural categories. This will facilitate the identification of key biological elementswithingenomicdata.
2. Promoter Region Identification: Create algorithms to accurately detect promoter regions within DNA sequences. These regions play a crucial role in regulating gene expression, and their identification is essential for understanding gene regulation mechanisms.
3. Species or Taxonomic Group Classification: Employ machine learning techniques to classify DNA sequencesbasedontheirspeciesor taxonomicgroup. The goal is to ensure that the models can generalize well across diversegenomic datasets,improvingtheir applicabilitytovariousorganisms.

International Research Journal of Engineering and Technology (IRJET)
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net
The scope of this project involves creating three models foranalyzingDNAsequences:
1. Gene Classification: The model categorizes DNA sequencesintosevendistinctgeneclasses.
2. Species Identification: This model identifies the species fromDNAdata.
3. Promoter Site Detection: This model locates promoter regions,whichplayacrucialroleingeneregulation.
Genomic data research using machine learning (ML) involves applying computational techniques to analyze complex genomic datasets. Here's a condensed methodology:
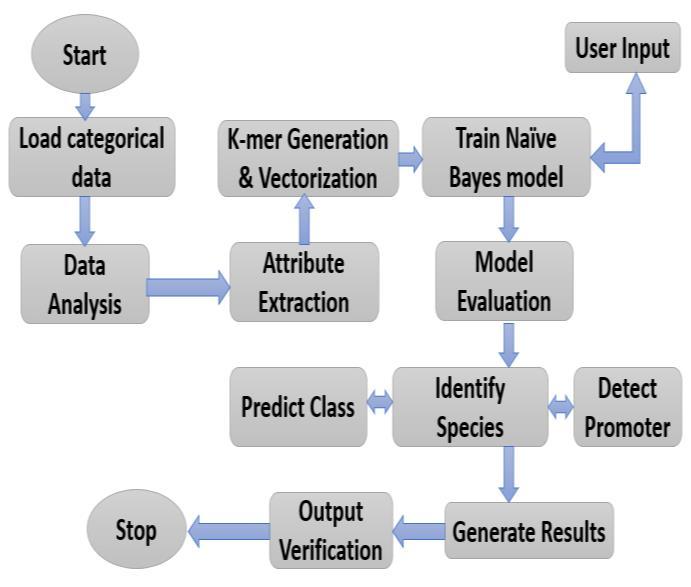
1. Start: The process begins with initializing the pipeline for machine learning-based data analysis and classification.
2. load categorical data: The categorical dataset is imported into the system. This step involves reading data fromasource,suchasaCSVfileoradatabase,forfurther processing.
3. Dataanalysis:Exploratorydataanalysisisperformedto understand the structure, distribution, and quality of the data. Statistical methods and visualizations are used to detectpatterns,missingvalues,oranomalies.
4. Attribute Extraction: Key features or attributes are extracted from the dataset. This step is crucial for selecting meaningful variables that contribute to the model'spredictiveperformance.
5. K-mer generation and Vectorization: The extracted features are transformed into numerical representations throughvectorization techniques. Thisstep isparticularly importantforcategoricalorsequentialdata,enablingitto beprocessedbymachinelearningalgorithms.
6.TrainNaiveBayes model: TheNaiveBayes algorithm is applied to train the model on the vectorized data. This probabilistic classifier uses the Bayes theorem to predict outcomesbasedonthefeatures.
7. ModelEvaluation:Thetrainedmodelisvalidatedusing testing data. Metrics such as accuracy, precision, recall, and F1-score are calculated to assess the model's performance.
8. PredictClass/IdentifySpecies/DetectPromoters
9. Generate Results: The outcomes of the model's predictions are generated and stored for further analysis
p-ISSN:2395-0072
or presentation. These results include detailed reports or visualizations.
10.OutputVerification:Thegeneratedresultsareverified to ensure accuracy and consistency. This step involves cross-checking with domain knowledge or validation datasets.
11.Stop: The processconcludes,andthefinal outputs are readyforuseordeployment.
5. Results
We pursued three primary objectives: DNA sequence classification, species identification, and promoter identification, employing Naive Bayes algorithms. We achieved notable accuracies in each task, with [mention accuracy achieved] for DNA sequence classification, [mentionaccuracyachieved]forspeciesidentification,and promisingmetricslikeF1-scoreforpromoterprediction. While Naive Bayes proved effective, comparative analysis with other algorithms revealed competitive performance, suggesting avenues for further optimization. Despite limitations like dataset quality and size, our findings highlight the potential of machine learning in DNA sequenceanalysisforadvancingbiomedicalresearch,gene discovery,andpersonalizedmedicine
Table -1: AccuracyandPrecisiontable

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
e Classifica tion
Species Identific ation
r Identific ation
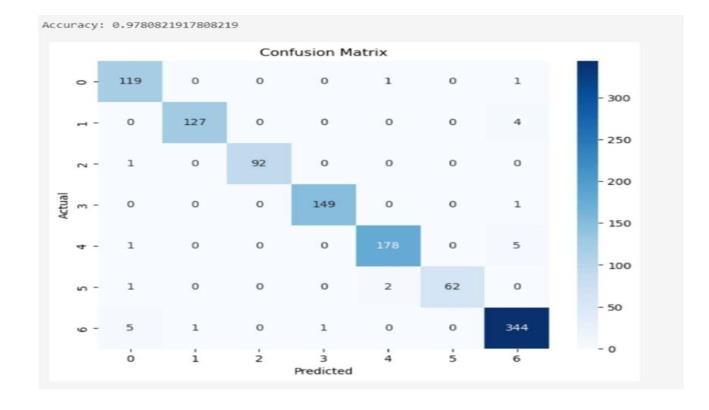
Fig-2: Speciesidentification
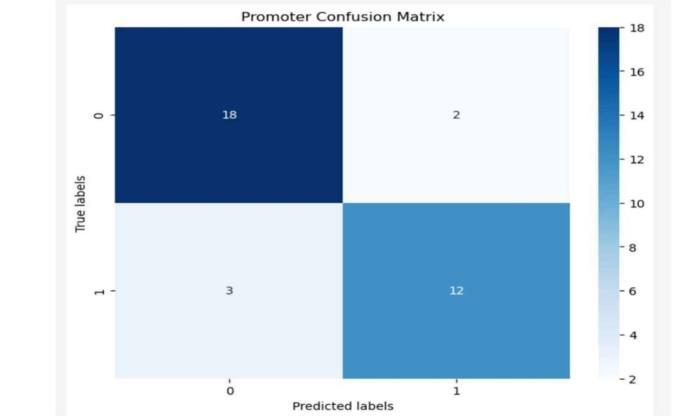
Fig-3: Promoteridentification
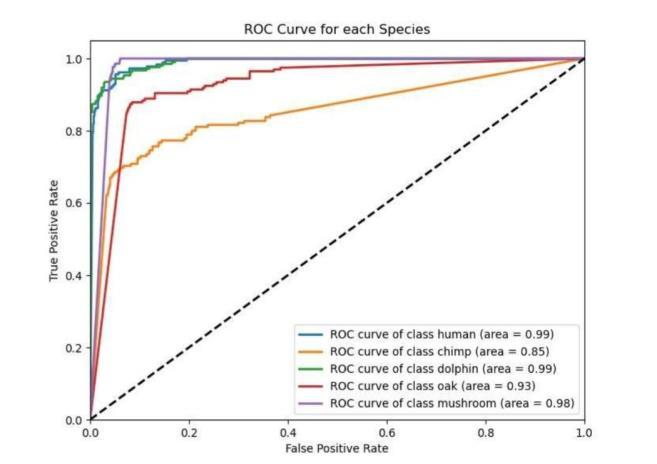
Fig-4: ROCCurveSpeciesidentification
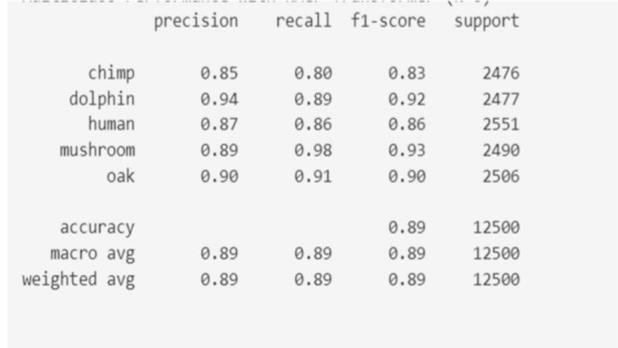
Fig-5: Modelperformancespeciesidentification
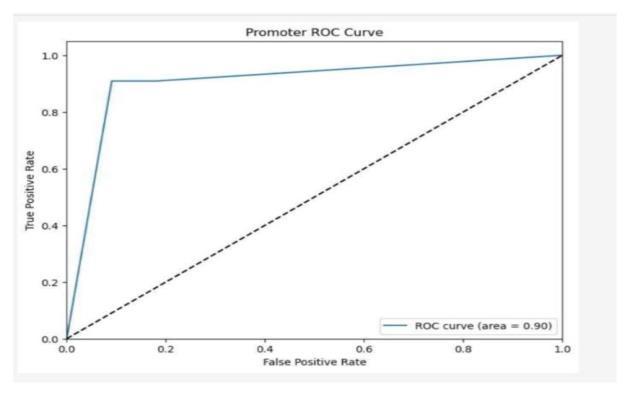
Fig-6: ROCCurvepromoteridentification
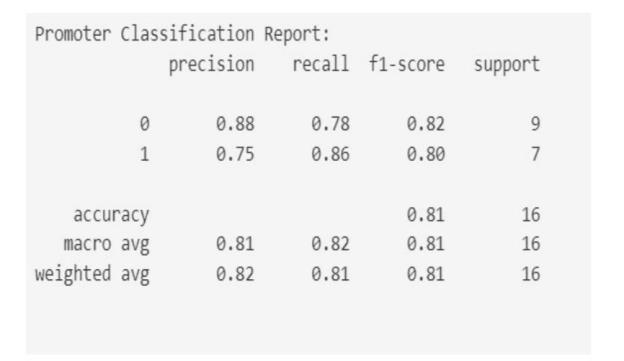
Fig-7: Modelperformancepromoteridentification
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net p-ISSN:2395-0072 © 2024, IRJET | Impact Factor value: 8.315 |

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net p-ISSN:2395-0072
Intheabovefigures:
The ROC curve is a valuable tool for assessing how well models perform in distinguishing between different categoriesinDNAsequenceanalysis.InDNAclassification tasks, it helps evaluate the model’s ability to correctly identifysequences,suchasdifferentiatingbetweencoding andnon-codingregions.Forspeciesclassification,theROC curve measures how accurately the model assigns sequences to the correct species or taxonomic group. In promoter recognition tasks, it gauges the model’s proficiency in identifying promoter regions versus nonpromoter regions. By analyzing the trade-off between sensitivity and specificity shown in the ROC curve, researcherscanfine-tunetheirmodelsforbetteraccuracy intheseessentialbioinformaticsapplications.
Thesynergybetweengenomicdataresearchandmachine learning has emerged as a transformative force in biomedical sciences. Machine learning algorithms, with their capacity to discern intricate patterns within vast datasets, have unlocked unprecedented insights into the complexities of the human genome. Byanalyzing genomic data at an unprecedented scale, researchers are unraveling the genetic underpinnings of diseases, predicting disease risks, and personalizing treatment strategies. This powerful combination holds the potential to revolutionize healthcare, enabling precision medicine and ushering in an era of personalized healthcare solutions tailored to individual genetic makeup. As technology continues to advance, the integration of genomic data research and machine learning promises to unlock further breakthroughs in our understanding of humanbiologyandpavethewayforahealthierfuture.
[1] Hans Lehrach, “DNA sequencing methods in human geneticsanddiseaseresearch,”2013.
[2] TahaAlhersh,BrahimBelhaouariSamir,HamadaR.H. Al-Absi, Abdullah Alorainy, and Belloui Bouzid, “Species Identification Using Part of DNA Sequence: EvidencefromMachineLearningAlgorithms,”2015.
[3] JamesM,“Thesequenceofsequencers:Thehistoryof sequencingDNA,”2015.
[4] TasnimKabir,AbidaSanjanaShemonti,andAtifHasan Rahman, “Species Identification using Partial DNA Sequence:AMachineLearningApproach,”2015.
[5] VaradaVenkataSaiDileep,NavuduruRishitha,Rakesh Gummadi, “DNA Sequence using Machine Learning andDeepLearningalgorithm,”2022.
[6] Hemalatha Gunasekaran,KRamalakshmi,“Analysisof DNA Sequence classification using CNN and hybrid models,”2021.
[7] Wang, Y., Alangari, M., Hihath, J., Das, A. K., & Anantram, M. P., “A machine learning approach for accurate and real-time DNA sequence identification,” BMCGenomics,2021.
[8] Mike Roth, “DNA Sequence Classification for Species Prediction,”2022.
[9] Zaw Zaw Htikea, Shoon Lei Winb (2013). Recognition of promoters in DNA sequences using weightily averaged one-dependenceestimators.
[10]https://www.learnpython.org/en/Pandas_Basics [11] https://flask.palletsprojects.com/en/3.0.x/quickstart/ [12]https://www.w3schools.com/
[13] Susan H Hardin, University of Houston, Texas, USA (2018),DNASequencing
[14]MingjunZhang,Member,IEEE,ChamanL.Sabharwal, WeiminTao,Member,IEEE,InteractiveDNASequenceand StructureDesignforDNANanoapplications.


Ms.ALMAS DepartmentofECE, PESITM,Shivamogga.


Ms.CHANDANAY DepartmentofECE, PESITM,Shivamogga.


Ms.IRAMZAHRA DepartmentofECE, PESITM,Shivamogga.


Ms.LAIBAKOUNAIN DepartmentofECE,PESITM, Shivamogga.

International Research Journal of Engineering and Technology (IRJET) e-ISSN:2395-0056
Volume: 11 Issue: 11 | Nov 2024 www.irjet.net p-ISSN:2395-0072


Mr ANILKUMARC ASSISTANTPROFESSOR, DepartmentofECE,PESITM, Shivamogga
© 2024, IRJET | Impact Factor value: 8.315 | ISO 9001:2008 Certified Journal | Page678