scıentific Carolina
SPRING 2024 | Volume 18 | Issue 2

—HOW DIABETES
AFFECTS
THE BRAIN— full story on page 18

SPRING 2024 | Volume 18 | Issue 2

—HOW DIABETES
THE BRAIN— full story on page 18





Check out all of our previous issues at issuu.com/uncsci. As the organization continues to grow, we would like to thank our Faculty Advisor, Dr. Lillian Zwemer, for her continued support and mentorship.












Mission Statement:
Founded in Spring 2008, Carolina Scientific serves to educate undergraduates by focusing on the exciting innovations in science and current research that are taking place at UNC-Chapel Hill. Carolina Scientific strives to provide a way for students to discover and express their knowledge of new scientific advances, to encourage students to explore and report on the latest scientific research at UNC-Chapel Hill, and to educate and inform readers while promoting interest in science and research.
Letter from the Editors:
As the weather warms and another semester at Carolina comes to a close, our school community can be proud of the academic and personal achievements that have come with it. Here at Carolina Scientific, we have continued to grow and foster more effective scientific communication through a Spring issue filled with topics ranging from ADHD research to neutrino physics. In addition, this semester has brought on a new avenue for growth with the release of a blog series; this series aims to make research even more accessible and approachable to the thousands of students here at UNC. The work done by Carolina Scientific would not be possible without the help of our writers, editors, and designers. As you flip through the pages of our work, we hope we’ve made science more fun and understandable.
- Sarah Giang & Isaac Hwang
Editors-in-Chiefs Sarah Giang
Isaac Hwang
Design Editor Cassie Wan
Copy Editor Corinne Drabenscott
Managing Editor Meitra Kazemi
Treasurer
Ambika Bhatt
Secretary Natalie Druffner
Publicity Chair
Heidi Cao
Fundraising Chair Hari Patel
Associate Editors
Esha Agarwal
Kruti Bhargav
Julia Boltz
Sprihaa Kolanukuduru
Andrew Phan
Jasmeet Singh
Online Content Manager Sreya Upputuri
Faculty Advisor Lillian Zwemer, Ph.D.
Staff Writers Copy Staff
Aiden Balandan
Ananya Solanki
Anish Aradhey
Angela Liu
Anooshka Deshpande
Ashley Hardner
Blake Seigler
Devin Glover
Dianne Celemen
Ellie Rogers
Gayatri Venkatesan
Grace Greenberg
John Wadington
Kaitlyn Campbell
Katharine Peck
Khadeejah Saleem
Kirina Shah
Lilly Baker
Masha Dixon
Matthew Rodzen
Medha Das
Megan Brantly
Morcos Saeed
Noha Goumaa
Oliver Ewy
Paige Twohill
Preston Szczesniak
Rahul Shah
Reem Fayyad
Ria Patel
Ruhi Saldanha
Samantha Breen
Sarah Edmonds
Shreeya More
Skye Scoggins
Srinidhi Subramaniam
Sophia Harvey
Valen Chapel
Zichen Zhang
Abigail Wells
Aditi Deshpande
Aiden Balandan
Akash Bhowmik
Akhil Malakapalli
Alacia McClary
Alexander Kinrade
Amelia Bruns
Ana Barton
Angela Liu
Arora Rohrbach
Arushi Vaish
Baylee Materia
Cindy Lam
Cyria Olingou
Daniela Danilova
Dhruv Garg
Gayatri Mehra
Gayatri Venkatesan
Julia Sallean
Keerthana Mariappan
Kelly Yun
Klodia Badal
Lillian Hohn
Madison Reavis
Mckenzie Miller
Michele Phipps
Nastia Hnatov
Natalie Druffner
Navya Maheshwari
Nicholas Boyer
Paige Strecker
Paige Twohill
Quinten Curtis
Rachna Sehgal
Raife Levy
Razmin Bari
Sabrina Kolls
Sai Satvik Kolla
Samara Williams
Sara Boburka
Sarah Edmonds
Sevilay Betul Coskun
Sophia Huang
Taylor O’Connor
Thomas Rempel
Tsehai Ricketts
Yasmine Ackall
Zaid Syed
Abigail Wells
Alejandra Ramirez
Arushi Vaish
Bhavika Chirumamilla
Caroline Norland
Cassie Wan
Cindy Lam
Clara Lord
Estella Calcaterra
Heidi Cao
Heidi Segars
Jacqueline Nguyen
Karen Zhu
Katrina Murch
Kelly Yun
Lorie Nguyen
Makayla Barnes
Matthew Rodzen
Noha Goumaa
Sara Boburka
Spoorthi Marada
Srinithi Gali
Sydney Rohm
Tanisha Choudhury
Sociology
Embracing Two Cultures
Dianne Celemen
Add Health
Grace Greenberg
Neutrinos Could Explain Why the Universe Exists?
Anish Aradhey
Tiny Technologies, Big Dreams
Blake Seigler
At the Crossroads of Mathematics and Medicine
Megan Brantly
A New Solution to an Old Problem
Ananya Solanki
How Diabetes Affects the Brain
Anooshka Deshpande
Calculator to Clinician
Ashley Hardner
Heart Failure Prevention May Be Just a Receptor
Away!
Kaitlyn Campbell
Splitting Sequences
Paige Twohill
The RNA Revolution in Cancer Treatment
Ruhi Saldanha
A Deep Dive into Precision Nutrition
Samantha Breen
Lymphoma in the context of HIV in Sub-Saharan
Africa
Srinidhi Subramaniam
Making Breakthroughs in the Biome Below
Sophia Harvey
Helping the Smallest Patients Fight a Large Battle
Valen Chapel
Autism’s Algorithm
Aiden Balandan
Flies Aren’t Just Winging It
Angela Liu
Laughter Isn’t the Best Medicine, Self-Compassion Is
John Wadington
Face Your Fears, or Don’t
Katharine Peck
Unraveling the Mysteries
Kirina Shah
The Magnificent Powers of Memory Modification
Medha Das
The ABCs of Infant Attention and ADHD Symptoms in Early Childhood
Noha Goumaa
Exploring the Connection of Addiction and Neurogenesis
Reem Fayyad
When Mom Drinks, Her Baby Drinks
Ria Patel
Unmasking the Pandemic’s Impact
Shreeya More
Skye Scoggins
Biology
Lipid Code
Devin Glover
Deciphering Gut Dynamics
Ellie Rogers
Metabolic Maestros
Gayatri Venkatesan
Dangers of Antibiotic Tolerance
Khadeejah Saleem
Leading Without a Nucleus
Lilly Baker
Everything Changes
Masha Dixon
Unraveling the Dance of Cells
Matthew Rodzen
Safeguarding Cardiac and Neuronal Cell Health in Space Travel and Aging
Morcos Saeed
Plant Immunity
Oliver Ewy
Plant Research and Conservation at the North
Carolina Botanical Gardens
Preston Szczesniak
Purifying Our Skies
Rahul Shah
Walking Lungs
Sarah Edmonds
Survival Through the Ages
Zichen Zhang

In a world where cultural boundaries blur and identities intertwine, the concept of biculturalism emerges as an increasingly relevant topic. Now more than ever before, the United States is witnessing unprecedented diversity characterized by individuals from multiple ethnic and racial backgrounds, a trend mirrored globally. Shifting demographics carry significant implications for both societal structures and individual development. Broadly, biculturalism is defined as internalizing two cultural systems, such as one’s heritage and national cultures, and developing the ability to navigate between these cultures.1 Thus, biculturalism is a multifaceted construct that involves many domains (e.g., values, practices) and strategies (e.g., codeswitching, integration). One important facet of biculturalism involves bicultural competence, which refers to the skills needed to meet the expectations of both one’s heritage and national cultures, especially among those from ethnic-racial minoritized backgrounds and immigrant communities. Bicultural competence includes 1) bicultural facility, or navigating bicultural expectations easily, and 2) bicultural comfort, or having positive emotions linked to managing bicultural expectations.2 Cultural and developmental scholars consider each of these facets when examining antecedents and consequents of biculturalism.
One of these scholars is Dr. M. Dalal Safa, a faculty member in the Department of Psychology and Neuroscience at The University of North Carolina at Chapel Hill (UNC-Chapel Hill), whose research focuses on biculturalism development
among adolescents and young adults. Leveraging her own experiences living and conducting research in several countries, Dr. Safa seeks to understand developmental trajectories of biculturalism and their links to positive adjustment. Extensive literature supports the positive association between biculturalism and adjustment across multiple settings;3 however, more research is needed to understand the mechanisms that promote or undermine biculturalism development. Dr. Safa’s program of research features longitudinal studies, measuring data at different time points and intervals throughout adolescence and young adulthood. She considers this longitudinal approach combined with qualitative and applied research to be features of the Biculturalism, Resilience & Identity Development in Global Environments & Systems (BRIDGES) Lab, a team of scholars she leads at UNCChapel Hill.1
The BRIDGES Lab is committed to elucidating cultural bridges that can support individual positive development in multicultural environments (Figure 1).


Figure 1. The BRIDGES Lab at UNC is committed to examining cultural bridges that can support positive development in multicultural environments and systems. Image courtesy of Dr. M. Dalal Safa.
Dr. Safa remarks, “We live in a world that is becoming more and more multicultural. The chances of encountering others who were raised differently than us are great; thus, studying and promoting cultural competencies is important. The research conducted in the BRIDGES Lab has key implications for the well-being of people living in multicultural societies, and it extends beyond the field of psychology.”1 Biculturalism research prompts, thus, a necessary discussion about developing and using cultural competencies in various settings — from personal relations to professional and academic settings. Given the BRIDGES Lab’s concentration on developmental trajectories, one focus of their work is on young adults transitioning from high school into college, an important transitional period. During this time, youth encounter experiences that may have lasting effects on their self-
perception and self-identification. Balancing one’s heritage and national cultures can influence youth adjustment in various social contexts, especially in academic settings.
In 2022, Dr. Safa explored the link between bicultural competence trajectories and academic adjustment among Latino/a/x youth transitioning from high school into college (data came from the Transiciones study led by Dr. Doane). Specifically, longitudinal trajectories of bicultural competence including bicultural facility and comfort were examined in relation to assessments of academic self-efficacy, academic engagement, and academic achievement (e.g., GPA scores).4 Findings indicated that youth trajectories of bicultural competence were found to simultaneously promote (e.g., positive trajectories associated with higher academic self-efficacy and engagement) and undermine youth’s academic adjustment, even within the same school context (Figure 2). It is possible that professors’ support of students’ bicultural skills and identities can increase the academic-related benefits of bicultural competence. Microaggressions from peers, however, can decrease the benefits that students draw from code-switching, which is a key behavioral, affective, and cognitive task enabling bicultural individuals to navigate between cultural perspectives from both heritage and national cultural systems.4

code-switching) during the adaptation to college.5 The study examined how trajectories of bicultural facility and comfort, accounting for the role of exposure to discrimination across time, influenced psychological well-being and substance use. First, findings indicated that greater experiences of discrimination undermined youth trajectories of bicultural competence. Further, longitudinal trajectories of bicultural facility and comfort did not predict substance use, but positive trajectories of bicultural facility were associated with better psychological well-being. Study findings prompt further investigation on the clinical applicability of the effects of biculturalism, such as in targeted interventions and healthy coping behaviors to college stressors. The study captured the interplay between stressors (i.e., discrimination) and cultural competencies (i.e., bicultural facility and comfort) in promoting or undermining adjustment throughout the transition to college.5
More recently, the BRIDGES Lab has been engaging in applied and interdisciplinary research. For instance, Dr. Safa has directly worked with school teachers given their crucial role in youth development, including the development of bicultural competencies and identities.1 To elucidate developmental mechanisms,6 the BRIDGES Lab is currently examining the impact of a school-based intervention on youth’s ability to find harmony and resolve conflicts across their heritage and national identities (My Life, My Identity Follow-Up longitudinal study in partnership with Drs. Sladek and UmañaTaylor). As Dr. Safa described her ongoing research projects,1 she highlighted the value of collaborative science and the invaluable contributions of her mentors, students, research partners, participants, and funders (i.e., NSF and SRCD).
media influences the development of biculturalism and its benefits for youth adjustment. Keeping youth at the center of each study is of utmost importance in creating supportive programs tailored to bicultural youth who navigate the complex dynamics between their heritage and national cultures.

Figure 3. Identities composed of multiple cultures are becoming increasingly prevalent in today’s global societies. Image courtesy of Wikimedia Commons.
Building on this work, doctoral student Michaela S. Gusman and Drs. Safa, Grimm, and Doane investigated how Latino/a/x youth’s exposure to discrimination shaped bicultural facility and comfort trajectories, as increased exposure to microaggressions might stunt these youth’s ability and confidence in using their bicultural skills (e.g.,
The high relevance of biculturalism research in today’s increasingly diverse society cannot be overstated. Disseminating these novel findings outside academic settings can prompt honest conversations about cultural competencies and multiculturalism. Dr. Safa remarks on her excitement to move towards a global society where people value multiculturalism and understand its benefits and challenges (Figure 3). In the future, the BRIDGES Lab anticipates exploring how the digital space of social
1. Interview with M. Dalal Safa, Ph.D. 02/15/2024.
2. Safa, M.D.; White, R.M.B.; Knight, G.P. Child Dev 2021, 96, 1211-1227.
3. Safa, M.D.; Umaña-Taylor, A.J. Adv. Child Dev. Behav. 2021, 61, 73-127.
4. Safa, M.D.; Gusman, M.S.; Doane, L.D. Child Dev. 2022, 93, 1663-1679.
5. Gusman, M.S.; Safa, M.D.; Grimm, K.J.; Doane, L.D. Clin. Psych. Sci. 2023, 12, 320-343.
6. Safa, M.D.; Long, Y.; Umaña-Taylor, A.J. Int. Journal Behav. Dev. 2023, 10.1177/01650254231218284

Embarking on a journey through time, Add Health tells the complex narrative of American health from adolescence to adulthood. The longitudinal study at the University of North Carolina at Chapel Hill not only traces the evolving health patterns of individuals but also unravels the intricate threads connecting schools, families, peers, neighborhoods, and other contexts of social life within the stark disparities shaping the nation’s health landscape.
In general, longitudinal studies can be expensive, time-consuming, and difficult. However, they often produce invaluable data for research, especially when conducted properly. One of the largest, most informative longitudinal surveys of adolescents who have now transitioned to midlife is The National Longitudinal Study of Adolescent to


Adult Health or Add Health, conducted at UNC-Chapel Hill. UNC-Chapel Hill faculty member Dr. J. Richard Udry initially directed Add Health, from 1994-2004. The second director of Add Health (20042021) was Dr. Kathleen Mullan Harris, who continues to contribute in important ways to Add Health. Since 2021, Dr. Robert Hummer, Howard W. Odum Distinguished Professor of Sociology and Faculty Fellow of the Carolina Population Center, has served as the director. Dr. Hummer received his Ph.D. from Florida State University and has spent his career particularly interested in studying US health/mortality trends, publishing his most recent book in 2019 with Erin R. Hamilton, entitled ‘Population Health in America’ (University of California Press).
content to the survey, which gave legs to the nickname ‘Add Health’ to the study. Add Health began in the 19941995 school year and has followed a nationally representative cohort of 20,745 adolescents in grades 7-12 across the country ever since. Participants were randomly chosen from a listing of eighty high schools across the nation. Given the study design, clusters of adolescents in grades 7-12 from the various schools were surveyed1. The survey’s original ability to track networks and study how friendships change and affect adolescents was a modern and innovative design. Data pertaining to groups from the same school provided insight into how different schools and backgrounds influence the lives of the adolescents involved. Diving into the details, the Wave I survey included questions about peer networks, sexual activity, substance use, health status, nutrition, family status, and more. Additionally, parents were encouraged to provide survey Dr.
Add Health was named initially from a survey of adolescents focusing on sexual behavior, drug use, and related topics associated with HIV/AIDS. The original study investigators were encouraged to add even more health

reports about the households in which the adolescents lived. Furthermore, biological data—such as height and weight—were recorded. Waves II through V of data collection included some of the same questions across time, as well as new content concerning educational transitions, economic status and strains, sleep health, physical activity levels, memory, biologically assessed health (e.g., blood pressure, cholesterol, etc.) as the adolescents made the transition to adulthood. So far, the data collected by Add Health has been used by over 50,000 researchers and has been included in tens of thousands of peer-reviewed journal articles, books, theses, dissertations, and research presentations on a wide array of topics. Research products have spanned from overall population health patterns to American adolescent social life, all of which have used data from this groundbreaking study.
Fast forward to 2024, and the Add Health team is in the field collecting data for Wave VI, largely funded by the National Institute on Aging. A large portion of the surveys are web-based, while some participants continue to answer the survey in-person. Wave VI includes selfreported data from the participants, as well as biological data collected using blood samples. The cohort is currently in their mid-40s and collecting data can be difficult. For instance, it is expensive to get out into the field and travel to individuals’ homes all over the country to collect biological samples. Also, achieving high response rates is tricky, and formulating strategies to increase response rates can be expensive. Some of the especially innovative data being collected in this wave include cognitive functioning, physical functioning, and hearing.1 One of the main goals of Wave VI is to develop a better understanding of adults’ functioning across their lives, given the challenges that this cohort has faced over the years, including the Great Recession, political polarization, the opioid epidemic, and COVID-19. Further, there is an emphasis on the mental
health of the cohort. Researchers have emphasized asking questions concerning mental health, complicated by whether they are caregivers, juggling careers and families, or just juggling the stresses of life as a middle-aged adult. The data for Wave VI should be available to researchers in mid-to-late 2025.
There are many reasons why Add Health is unique in what it brings to the table of population health studies, but Dr. Hummer stated three of the most important.1 The Add Health birth cohort, born around 1980, is the first cohort to live their full lives in an ‘obesogenic environment’. The obesogenic environment refers to an environment that promotes weight gain through things such as easily accessible unhealthy foods, and daily activities that do not include exercising. Add Health has highlighted that the levels of diabetes and hypertension, as well as markers of these impending conditions, were concerningly high as early as Wave IV. At that time, the average age was only 28 years old for this cohort. This was a key pattern identified by Add Health when examining the health of the average American young adult today. Another key finding of Add Health is how important school and family connections and relationships are for the overall health of individuals. Those findings were somewhat known previously but concretely demonstrated by publications written using Add Health data. A final key innovation that Dr. Hummer highlights is the focus of Add Health on inequality. Dr. Hummer says, “Add Health data continues to be really important nationally for understanding the development and maintenance of health disparities.” Overall, the study has the potential to create a better understanding of ethnoracial health disparities and what social, economic, and policy contexts have to do with these disparities.2 Indeed, Add Health researchers have clearly shown that adolescence is an important point in the life course when health disparities truly begin to emerge, which is an interesting
point for further investigation as the cohort continues to age.
Long-term, Dr. Hummer and his colleagues plan to continue collecting data on the Add Health cohort as they age into older adulthood. While it will be tremendously challenging to pull this off, it will be more than worth it, as this study and the data it has been producing for the last few decades is invaluable. In the words of Dr. Robert Hummer: “In the end, if we’re successful, we can tell the American Story, which is priceless.”
1. Interview with A. Robert Hummer, Ph.D. 02/14/2024
2. Hummer, R. A. Race and Ethnicity, Racism, and Population Health in the United States: The Straightforward, the Complex, Innovations, and the Future. Demography 2023, 60 (3), 633–657. https://doi.org/10.1215/0070337010747542.
Physicists know the Big Bang created equal amounts of matter and antimatter. But nearly everything we observe today, including ourselves, is matter. Physicists seek an explanation for our matter-dominated universe from a tiny particle which they have named the neutrino.
Antimatter particles have the same mass but opposite electrical charge than their matter counterparts. For example, a positron—the antiparticle of an electron—has a positive charge. According to Einstein’s famous equation E=mc2, when antimatter and matter collide, they transform each other into pure energy. Therefore, the equal antimatter and matter in the early universe should have destroyed each other, leaving no mass behind. However, judging from the matter-rich universe we live in today, this did not happen. “Somewhere, there is an imbalance in the early universe that creates slightly more matter than antimatter,” said Dr. John Wilkerson, a neutrino researcher and Distinguished Professor in the Department of Physics and Astronomy at the University of North Carolina at Chapel Hill.1 Physicists believe that this imbalance could occur if neutrinos behave as their own antiparticles. Nearly a century ago, Italian physicist Ettore Majorana (my-oh-RAW-nuh) first posed this question. Today, physicists like Dr. Wilkerson are closer than ever to finding an answer.

Fundamental particles that cannot be divided further are the building blocks of the universe. Of these particles, the ghostliest is the neutrino: a tiny particle that slips straight through most matter. About 100 billion neutrinos zoom through your thumbnail every second.2 However, unlike other small particles—such as electrons and protons, whose mass and charge cause them to interact with nearby particles—neutrinos’ extremely low mass and lack of charge means they barely interact with their surroundings. This makes them extremely hard to detect. Although scientists do not know the exact mass of a neutrino, the large KArlsruhe TRItium Neutrino (KATRIN) experiment in Germany recently discovered that neutrinos must be at least half a million times lighter
than an electron.3

Physicists first theorized the existence of these ghost particles over 100 years ago when studying radioactive materials that undergo a shape-shifting process called beta decay. During beta decay, one neutron in the atom’s nucleus spontaneously transforms into a proton and an emitted electron. For example, Carbon 14 (14C), which is used to estimate the age of archaeological finds, decays into Nitrogen 14 (14N).4 When physicists first observed this decay, the energy and momentum of the emitted electron seemed to violate the laws conserving energy and momentum. Therefore, they predicted that beta decay must also emit another, extremely light particle. It would take physicists another 25 years to detect neutrinos.
Dr. Wilkerson is one of several UNC-Chapel Hill researchers who are involved with international scientific collaborations that aim to discover whether neutrinos are their own antiparticle. These huge experiments focus on detecting a never-seen-before decay: neutrinoless double beta decay. In the nuclei of materials such as the lustrous metal germanium 76 (76Ge), there is a chance of two neutrons simultaneously morphing into two protons and emitting two electrons and two neutrinos. If neutrinos are their own antiparticles, the two neutrinos should annihilate each other into energy (E=mc2), making the process “neutrinoless.” “These experiments are conceptually very simple,” Dr. Wilkerson explained. First, researchers create detectors from two to three kilograms of germanium, which function as both sensors and sources of double beta decay. They shroud these detectors with a curtain of sensor fibers and immerse this setup in a tank of frigid liquid argon (at –303°F, or –186°C). Together, the liquid argon and fibers can measure faint radiation from decays in the germanium at precise certain energy levels. “We know exactly where [the energy level] should be from mass energy conservation,” Dr. Wilkerson explained. Essentially, the experiments surround germanium with sensors and wait for a pair of neutrons to transform.
But these experiments are much harder to execute than it may seem at first glance. Firstly, neutrinoless double beta decay is an extremely rare process. The half-life of a material is defined as the time it would take for half of its nuclei to undergo a radioactive decay process; the materials capable of neutrinoless double beta decay have half-lives trillions of times longer than the age of the universe. To increase their likelihood of observing a decay, scientists must assemble enormous amounts of germanium. Even harder is the task of shielding the sensors from sources of background radiation that could drown out the radiation signature of a double beta decay.
Dr. Wilkerson compares this to standing on the field of Kenan Memorial Stadium during a home football game. Under the beaming stadium lights, would you be able to tell whether your friend, sitting in the stands on the other side of the stadium, had her phone screen lit up? “It’s very hard to pick that signal out if you’re blinded by all the other light,” he said. For this reason, researchers hunting these rare decays go to “extraordinary lengths” to reduce the surrounding noise. These lengths include building the experiments deep underground to protect them from cosmic rays, surrounding detectors with tanks of water to identify muons (energetic particles that can pass through even a kilometer of bedrock), and ensuring that the materials in and around the experiment will emit no radiation on their own. Instead of normal rock and dust, which contain minute amounts of radioactive isotopes, researchers surround their experiments with specially made copper and custom “clean” materials.
Using these strategies, UNC researchers have contributed to several experiments. UNC-Chapel Hill was the lead institution on the Majorana Demonstrator Collaboration, an experiment containing 30 kilograms of germanium at the Sanford Underground Research Facility in South Dakota. The experiment, which concluded in March 2021, set a half-life on neutrinoless double beta decay of > 8.3 × 1025 years, successfully detected two-neutrino double beta decay, and served as a proof of concept for larger experiments.5

even bigger. Dr. Wilkerson is currently helping develop LEGEND-1000, which will house a full metric ton of germanium (1000 kilograms, or around 2,200 pounds) in Italy. Over a decade in the making, LEGEND-1000 will require researchers to build sensors 100 times more sensitive and equipment 20 times cleaner than what is currently available. But scientists are up for the challenge. Over 50 institutions and 250 collaborators from over 10 counties have united to make this experiment possible.6 With one metric ton of germanium, this experiment expects to detect about 3-5 double beta decays after running for 10 years.

Figure 3. The LEGEND-200 experiment. The metal cylinders are detectors enriched with germanium, and the green tank in the background is filled with liquid argon. Image courtesy of Dr. Wilkerson.
If these researchers manage to observe neutrinoless double beta decay, they will confirm a 100-year-old hypothesis and could explain why we are here. Furthermore, learning more about the neutrino properties, including their mass, is an important way to test astrophysicists’ current theories of the universe’s formation and expansion. For Dr. Wilkerson, there is joy in the hands-on work this experiment demands: building physical experiments from scratch and troubleshooting technical problems. There is also joy in learning how the nature of these ghost particles impacts our universe. “The exciting thing is discovering something about nature that no one’s ever known before,” he said.
During the COVID-19 pandemic, UNC-Chapel Hill researchers and students helped build the Large Enriched Germanium Experiment for Neutrinoless ββ Decay (LEGEND) 200, which will contain 200 kilograms of germanium at the Laboratori Nazionali del Gran Sasso in Italy. “We have a laboratory in the basement [of Phillips Hall] where we actually do testing of these detectors,” Dr. Wilkerson said. The next step is to go
1. Interview with John F. Wilkerson, Ph.D. 02/09/2024.
2. Fermilab. Solar Neutrinos. https://neutrinos.fnal.gov/ sources/solar-neutrinos/ (accessed February 23rd, 2024).
3. The KATRIN Collaboration. Nature Physics 2022, 18, 160166.
4. U.S. National Oceanic and Atmospheric Administration. The Technical Details: Radioactive Decay. https://gml.noaa. gov/ccgg/isotopes/decay.html (Accessed February 23rd, 2024).
5. Sanford Underground Research Facility. Majorana Demonstrator. https://sanfordlab.org/experiment/majorana-demonstrator (Accessed February 23rd, 2024).
6. Radford, D. C. LEGEND-1000 Technical Update. Nuclear Science Advisory Committee, U.S. Department of Energy 2021
Imagine a world in which medical testing takes minutes instead of days, where the average cost of tests can be measured in cents instead of dollars.1 This is the amazing world of microfluidics, where lab equipment that usually costs hundreds of thousands of dollars can now be created for a few thousand.2 Achieving this world is the goal of the Ramsey group and its principal investigator Professor Michael Ramsey.
Starting his career at Oak Ridge National Laboratory (ORNL) with a Ph.D. in Analytical Chemistry from Indiana University, Dr. Ramsey began by investigating laser-based chemical measurement technologies. However, when a small detour into the world of microfluidic technologies generated overwhelmingly positive feedback from companies and researchers alike, he decided microfluidics was where he should turn his focus. The emerging field of microfluidics studies fluids traveling in microscopic channels which can be used to coordinate and perform a variety of biochemical reactions such as those vital to the medical diagnostics industry. With this new excitement surrounding his work, Dr. Ramsey began to reprioritize his lab and build up a strong team to capitalize on the opportunity. After growing his team at ORNL however, he decided he needed a change of scenery and brought his team to the University of North Carolina at Chapel Hill as the Minnie N. Goldby Distinguished Professor of Chemistry, where he started the Ramsey Group in the Department of Chemistry and continued his miniaturization efforts.

The Ramsey Group’s research can be divided into 4 categories: Microfluidic Point-of-Care (POC) and High-throughput Technologies (HT), Microchip electrophoresis-
electrospray devices, Microscale Mass Spectrometry (MS), and Nanofluidics.3 Microfluidic POC and HT technologies are focused on using microfluidics to develop faster and less expensive tools to access biological information relevant to disease diagnosis and therapeutics development. One of the biggest problems that the group is trying to solve in this area involves developing precision biochemical assays to reduce time to results, provide more sensitive and precise measurements, while also reducing their costs.

Figure 1. Plastic microfluidic chip capable of performing multiplexed digital PCR assays to identify pathogens in samples such as blood or saliva. Photo courtesy of Professor Ramsey.
The second area of research is microchip electrophoresiselectrospray devices, which are used to separate ions in solution by applying a high voltage to a liquid. The liquid-phase ions are then transported to the gas phase similarly using a strong electric field.4 The gas-phase ions can then be characterized in detail using mass spectrometry.
The Ramsey Group extended their adventures into miniaturization of chemical measurement technologies by investigating the possibilities of microscale mass spectrometry. Conventional mass spectrometers are large and expensive machines that can weigh hundreds of pounds and cost hundreds of thousands to millions of dollars. By miniaturizing this technology these devices can become cheaper and more accessible, even to the point of being small and cost-effective enough for handheld instruments for identification of materials

Figure 2. A microscale ion trap mass analyzer patented by the Ramsey Group and used in the handheld mass spectrometers sold by 908 Devices. It is over 100x smaller than conventional devices. Photo courtesy of Professor Ramsey.
in the field. The fourth area of research is focused on nanofluidics. Nanofluidics are simply microfluidics done on the nanometer scale rather than the micrometer scale. This reduction in size allows the precise manipulation of single molecules, such as DNA, RNA, and proteins. The Ramsey group has worked on nanofluidic devices for characterizing long range genetic variations in single DNA molecules. Such genetic variations are generally inaccessible using present day DNA sequencing strategies but are believed to have important implications in many disease states.
Dr. Ramsey is not only a member of the Chemistry Department, however. He is also a professor of Biomedical Engineering and as of 2014, he is one of the founding professors of the Department of Applied Physical Sciences. As Professor Ramsey explains, “My primary appointment is [in] the chemistry department but most of our work has an engineering side to it…we’re very cross-disciplinary.”5 To show off the applicability of his research, Dr. Ramsey has spun out four companies from his laboratory during his career.
“My first company was spun out of ORNL: Caliper Technologies,” he explains. “They did an IPO in 1999.”5 Caliper Technologies was the first company to commercialize microfluidics technologies and primarily focused on life sciences tools applications, tools used by biotechnology and pharmaceutical companies. Caliper Technologies was later renamed Caliper Life Sciences and acquired by PerkinElmer in 2011. By then, Dr. Ramsey had already stepped away to join UNC-Chapel Hill.
He continues, “908 Devices was my second company, so that spun out from UNC, although the seminal IP was generated at ORNL again…We did an IPO in 2021.”5 908 Devices was founded with the goal of developing handheld mass spectrometry and miniaturized separation products that were originally developed in Dr. Ramsey’s lab.
The final two companies that the Ramsey Group has fostered are much newer. Genturi spun out of UNC and was founded in 2015 specializing in commercializing nanofluidics for long-range genetic variation testing in DNA. Codetta, the latest company, on the other hand was founded in 2021 and is focused on designing digital multiomics assay platforms for high throughput screening of biological targets related to disease diagnosis and therapeutics development.
With so many companies being spun out, Dr. Ramsey has shown himself to be adept at taking research and using it to make an impact. “My group’s research is in the area of applied sciences rather than basic research, and I feel like we need to do more than just write papers. So, starting companies and that produce products – it’s a way to give back to society.”5 With so many exciting projects in the works, Dr. Ramsey decided it was
best to change his scenery once again. “I’m much more involved with Codetta, obviously because of the proximity to Chapel Hill, and to a lesser extent still involved with 908 Devices. I also retired from UNC last week, so I have more time now to spend with the commercial side of things.”5
Even though Dr. Ramsey has decided to take a step back from microfluidics research, the field still races on. With so many opportunities in the biomedical industry calling for less expensive and more efficient access to biochemical information, it is hard to find many fields that will be more revolutionary for medicine and chemistry.

Figure 3. A cassette holding four microfluidic chips for digital nucleic acid or protein assays. Designed for high throughput, each chip can process approximately 2 million assays each. Photo courtesy of Professor Ramsey.
1. Ha, N. S.; De Raad, M.; Han, L. Z.; Golini, A.; Kim, Y.; Northen, T. RSC Chemical Biology 2021, 2 (5), 1331–1351.
2. Nguyen, H.; Thach, H.; Roy, E.; Huynh, K.; Perrault, C. M. Micromachines 2018, 9(9), 461. --3
3. Ramsey Research Group. (n.d.). https://ramsey.chem.unc. edu/ (accessed March 18th, 2024).
4. Electrospray ionization. https://en.wikipedia.org/wiki/ Electrospray_ionization (accessed March 18th, 2024).
5. Interview with Michael Ramsey, Ph.D. 2/05/2024

In spring 2020, as the rest of the world shut down, researchers at the University of North Carolina at Chapel Hill (UNC) jumped into action to fight COVID-19. One of these researchers was Dr. M. Gregory Forest, who utilized his background in mathematics to illustrate how the immune system fights off the COVID virus within the respiratory tract.
At his core, Dr. Forest is a mathematician, but his adaptability and love for learning made his background applicable to numerous fields. After earning his Ph.D. in mathematics from the University of Arizona, Dr. Forest was on the faculty at Ohio State before moving to UNC in 1996 to develop an applied math program within UNC’s Department of Mathematics. He suddenly found himself surrounded by health research, and even with no formal background in medicine, he took part in these studies. “Over the past 27 years that I’ve been here” he said, “we have been approached, and we have approached, people all over campus. And the collaborations just mushroomed.”1

One of Dr. Forest’s largest collaborations is with the Marsico Lung Institute, where he combines foundational knowledge of the respiratory system with his own
background to produce mathematical models of lung biology and disease. These projects uncover pathologies of diseases like asthma, cystic fibrosis, and in particular, COVID-19.
For Dr. Forest, the rise of COVID-19 presented a natural extension of over two decades’ work with the Marsico Institute, as well as ten years working with Dr. Sam Lai in the UNC School of Pharmacy on viruses in mucus membranes. Since January 2020, Dr. Forest’s group has focused on mathematical modeling of respiratory infections within human hosts. One model quantifies the progression of the COVID-19 virus by analyzing the timing and intensity of the innate immune response throughout the respiratory tract.1,2
To start an infection, the virus binds to a healthy cell’s surface and implants its RNA, hijacking the cell’s machinery to manufacture RNA copies of itself that spread infection once released (Figure 1).1,3 The innate immune response is the body’s first mechanism fighting this process. During the innate response, infected cells secrete interferons—small molecules with three unique functions. They (1) signal neighboring cells to slow down or stop surface protein binding, temporarily making them immune to the virus, (2) stimulate immune cells known as macrophages to engulf infected cells or viruses, and (3) interrupt the infected cell’s life cycle, decreasing viral RNA release.1,2
The interferons’ impact on the COVID-19 infection is the focus of one of Dr. Forest’s models. The mathematical
relationships are not a new discovery: “Did we invent the math?”, Dr. Forest said. “No. Modeling chemical reactions has been around a long time, and in this case it’s just cell spikes and binding events that either lead to an infection or not.”1

Once the chemical reaction-based model was created, the task at hand was adapting it specifically for COVID-19 infections. The Forest team’s model uses two main source and sink functions, or equations representing overall growth and death rates of infected cells. The source and sink functions analyze concentrations of viruses and interferons predicted by partial differential equations, plus a probability rule describing cells switching between uninfected and infected states. The model inputs parameters corresponding to the three ways interferons curb infection, as well as mathematical depictions of viral load and infected cell behavior, fine-tuning the overall source and sink functions.2
Early attempts at modeling
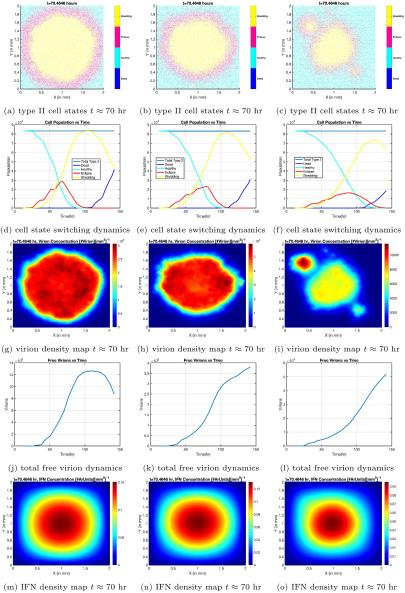
Figure 2. Cell phase distributions in space and time, virus concentrations vs. time, and interferon distributions in space. Each column corresponds to a different level of interferon-induced disruption in the infected cell’s life cycle, with the leftmost column having the lowest degree of disruption, and the rightmost column having the highest. Image courtesy of Dr. Forest.
the infection required more changes, however. The group’s first COVID models were agent-based, meaning each virus and cell was considered its own entity in a simulation that spanned the entire respiratory tract. According to Dr. Forest, this created a major problem with data analysis. “Just counting the number of viruses that are being generated in an infection overwhelmed our computers and storage capabilities”1 he said. This forced them to adopt a hybrid model, which represents viruses and interferons as concentrations—the number of particles per unit of mucus volume— rather than individual particles. This can only be done once the infection has sufficiently spread, and there are enough “agents” for the model to be scaled-up.
The novelty of the virus and vast differences in individual immune systems makes defining parameter values challenging for COVID researchers. In fact, some estimates fell in ranges varying up to 100 times the average value.1 For example, people have different rates of interferon release, thickness of mucus in the respiratory tract, and other unique
characteristics, which may impact infection progression in an individual. This became a major testable factor in Dr. Forest’s models; they easily redefine these parameters to account for unique immune systems. “This is where math comes in” Dr. Forest stated. “We perform a sensitivity analysis that reveals the degrees to which these measurable parameters have an outcome [on infection progression].”1
When run with complex numerical methods and algorithms, the model predicts the progression of viruses, interferons, and infected cells over time. Results showed the infection still substantially progressed, even with parameters tuned to represent high levels of interferon-induced immunity (Figure 2).2 Dr. Forest’s team showed the interferon-induced immune response is not enough to curb COVID-19 infection; as a result, early intervention after a positive test using medications like antibody treatments is vital, especially in those that are high risk.1,2
In-host mathematical models of COVID-19 have valuable real-world implications. Mathematical modeling expands a limited knowledge base surrounding dynamic progression of lung infections. In fact, within-host infection data is extremely limited: “You can’t go into someone’s respiratory tract and measure things in any level of detail” Dr. Forest commented. “It’s dangerous and unethical.”1 Some use cell culture or animal studies to help bridge this gap, but the Forest group’s mathematical models allow for a deeper analysis of these patterns with parameters specific to human anatomy and each person’s susceptibility to the virus.
Mathematical modeling also uncovers facts previously unknown to medical professionals; model results showed certain parameters have major impacts. For example, the time between successful virus-cell binding and the onset of RNA shedding exponentially impacts infection progression (Figure 3).1,3 Furthermore, the mathematical model accounts for the fact that mucus constantly moves in the respiratory tract to expel inhaled particles, accelerating viral load and infection. Because traditional experimental models—cell cultures—do not capture this motion, their viral load predictions vastly underestimate within-host values. This result was counterintuitive to colleagues
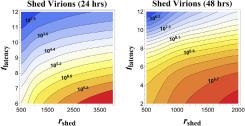
Figure 3. Number of viruses released by an infected cell at different release rates and lengths of time between the beginning of infection and the start of viral RNA shedding by the infected cell. When this time period decreases, and RNA release rate is held constant, the number of viruses increases substantially. Image courtesy of Dr. Forest.
at the Marsico Institute, as Dr. Forest points out: “How would they have known if we didn’t do the math?”1
In future studies, Dr. Forest hopes to include the effects of other aspects of the innate immune system such as T Cells for a more complete picture.1 Research into this field has come a long way already, however. “We know things about respiratory infections that we never knew before, just because there was a pandemic,” he said, “and it is amazing that this competition between viruses and the immune system… had not been modeled with spatial resolution faithful to human physiology previously for flu, cold, and other respiratory viruses.”1 In fact, the same algorithms and virus-specific parameters in the Forest group’s model can be adapted to model viral infections elsewhere in the body.1,2 Collaborations between two seemingly unrelated fields—mathematics and medicine— furthered understanding of a virus that turned the world upside down, and similar techniques will be used to study viruses still to come.
1. Interview with M. Gregory Forest, Ph.D., 02/09/24.
2. Aristotelous, A.C., Chen, A., Forest, M.G. J. Theor. Biol 2022, 555, 111293.
3. Pearson, J., Wessler, T., Chen, A., Boucher, R.C., Freeman, R., Lai, S.K., Pickles, R., Forest, M.G. J. Theor. Biol. 2023, 565, 111470.
The opioid problem has been around for centuries, and it has worsened within the past few decades. The opioid epidemic is unique because you cannot solve it by taking away opioids—the pain will not magically disappear. As long as there is pain, there must be a pain-relieving technique. But according to Dr. Grégory Scherrer, that technique may not have to be opioids.
Dr. Scherrer earned his PharmD and PhD from the University of Strasbourg in France, where he interned in the Oncology department. There, he was exposed to many who suffered from pain and were not always well served by the limited treatments available to them. This experience was the catalyst for focusing his graduate and post-doctoral studies on opioids and pain, leading him to join the lab of Dr. Brigitte Kieffer, the woman credited with discovering the gene that codes for opioid receptors.
Scherrer conducted a research experiment to identify the parts of the brain that make pain unpleasant. Pain is both a sensory and an emotional experience, yet little was known about the mechanism by which the information picked up by primary afferent nociceptors—the sensory neurons detecting and transmitting the sensation of pain to the brain—actually turned into the unpleasant emotional “feeling” of pain.1 In order to identify which neurons cause pain unpleasantness, the researchers had to find a way to observe specific neurons from the approximately 100 billion neurons present in the brain. Neurons work in highly interconnected networks, but each of them performs specialized functions. Therefore, in order to study them, the specialized cells needed must be isolated using sophisticated and inventive techniques. Dr. Scherrer explains that “oftentimes, the technology to do exactly what we need isn’t available, so we have to do it ourselves.”2

Dr. Scherrer’s lab focuses on understanding pain, opioid drugs, and their relationship to each other. The lab’s goal is to create new drugs for pain relief to alleviate doctors’ current reliance on opioids to help their patients (Fig 1). By targeting specific receptors, these researchers aim to create drugs that relieve pain without causing the negative effects associated with opioids, such as addiction, tolerance, or respiratory depression, which is the primary cause of overdose death.
"Pain can be difficult to treat with opioids over a lengthened period because heightening tolerance to the drug causes dose escalation, which can ultimately lead to addiction as a side effect."
In this experiment, Dr. Scherrer used a miniature microscope designed by his collaborator Dr. Schnitzer at Stanford, which could be mounted on a mouse head to view neural activation in a mouse’s brain upon pain sensation (Fig 2).2 A fluorescent protein was added to the neurons of the amygdala that activated when these cells fired. In past studies, Dr. Grégory Scherrer
In 2019, Dr.

Figure 2. With the miniature microscope attached to the mouse head, researchers could observe the areas of the mouse brain associated with the emotional response to pain. Image courtesy of Dr. Scherrer.
the basolateral amygdala (BLA) has been associated with many emotions, including those linked with pain perception. In addition, it has also been shown that damage to the BLA can remove the unpleasantness of pain while still allowing the sensation to be localized to different areas of the body.1 If the specific neurons that create negative pain perception could be isolated, then these neurons could be targeted for potential new drugs. Therefore, this experiment aimed to further understand exactly how the BLA encoded emotional distress in response to pain. When the mice were exposed to a physical pain stimulus, such as a needle prick, scientists could observe the precise nerve cells that lit up in the amygdala (Fig 3).2 To ensure that only the neurons associated with the emotional pain experience were isolated, other aversive but not painful stimuli were tested as well. Some examples include bitter taste, a loud noise, and a facial air puff. From these tests, a group of neurons was identified that only responded to the painful physical stimuli.2
The researchers realized that heightened activation of this group of neurons could be used to predict escalated pain

Figure 3. In addition to pinpricks for pain stimulus, heat and cold were used. The stimulation of neurons during light touch was also recorded, and a background is provided as a control. Image courtesy of Dr. Scherrer.
behaviors, implying that the processing of pain signals in the BLA influences the unpleasantness of the pain experience. While BLA activity is not responsible for the entire pain experience, it was concluded that the BLA could encode some negative emotions associated with pain and transmit this information to brain regions like the central amygdala or the striatum. This information is key to finding treatments for chronic pain, which is challenging to effectively solve long-term. Treatment with opioids over a lengthened period heightens tolerance to the drug, causing dose escalation, which can lead to addiction. This chain of events ultimately contributes to the opioid epidemic.
The end goal of Dr. Scherrer’s work is to solve this growing problem by formulating treatments that can be applied to patients in need. Currently, he and his lab are working with medical chemist Dr. Jeffrey Aubé at the University of North Carolina at Chapel Hill School of Pharmacy to develop new therapeutic drugs for pain. With the support of the University of North Carolina at Chapel Hill Eshelman Innovation, these researchers are devising new drugs to “turn off” pain perception pathways in the BLA. Through single cell RNA-sequencing, they have learned what genes were expressed in targeted cells of the amygdala. In particular, they found that many G proteincoupled receptors (GPCRs) produced in these cells could be potential targets of this new drug to curb nociceptive effects. As a part of the development process, different possible drugs were identified and tested against GPCRs for their efficacy in alleviating pain. A successful drug not only reduces pain, but is also safe and avoids causing harmful side effects like the addiction or respiratory depression caused by opioids. Results from early stages of experimentation have been positive so far, as the mice given these new treatments have experienced pain relief.1
Opioids can be very useful for some patients, but only so much can be done before their negative effects outweigh their good. As a consequence, opioids are used carefully, so pain is sometimes viewed as an inevitable outcome of certain medical conditions or procedures. But with the Scherrer lab’s work, that story can transition to a thing of the past. Dr. Scherrer and his colleagues’ discoveries about pain perception and work on innovative drugs are one of the many strides on the journey to solving the opioid crisis.
References
1. Corder G.; Ahanonu B.; Grewe B.F.; Wang D.; Schnitzer M.J.; Scherrer G. An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 2019, 363, 276-281.
2. Interview with Grégory Scherrer, P.h.D. 02/22/24

Over a billion people worldwide suffer from obesity.1 Individuals with obesity are at risk for a variety of conditions including diabetes because they have excessive adipose tissue, which can secrete hormones that interfere with metabolism [Figure 1]. Diabetes is a metabolic disorder in which blood glucose levels remain elevated, damaging vital organs such as the heart, kidneys, blood vessels, and eyes. Little work has been done to identify how the brain is impacted by obesity and diabetes. Dr. Janice Hwang, an endocrinologist and associate professor of medicine at the


Figure 1. Adipose tissue consists of cells called adipocytes which store fat and energy. It is primarily found underneath the skin, in bone marrow, and between organs. It also secretes hormones that impact metabolism. Photo courtesy of Wikimedia Commons
University of North Carolina at Chapel Hill, investigates the effects of obesity and diabetes on brain function to improve patient outcomes.
After a meal, blood glucose levels tend to rise. As a result, the pancreas produces insulin, a hormone that lowers blood glucose levels by removing glucose from the bloodstream and enabling it to enter cells [Figure 2]. Type 1 diabetes occurs when the pancreas produces little
to no insulin due to the autoimmune destruction of cells that produce insulin. Type 2 diabetes occurs because the body becomes less sensitive to insulin’s effects. This is often due to the impact of obesity. As a result, blood glucose levels remain high in both Type 1 and Type 2 diabetes. However, the treatment methods can vary significantly. Scientists have known for decades that obesity and diabetes have adverse
effects on the brain. A majority of this work revolved around animal and in vitro studies because it is very challenging to investigate the human brain. Recently, however, novel techniques have been developed to allow scientists to study the living human brain. Neuroimaging techniques such as magnetic resonance imaging can be very powerful tools. Scientists use a technique called magnetic resonance spectroscopy (MRS) to measure the concentrations of metabolites in the brain. In this technique, a magnetic field is applied to the patient’s brain, which causes the nuclei in the brain’s metabolite to spin and generate signals. By observing the behavior of the nuclei under the magnetic field, scientists can discern the concentration of certain
“Dr. Hwang believes that reduced glucose entry into the brain in type II diabetic patients could be a compensatory mechanism that protects the brain from high blood glucose levels.”
metabolites in the brain.
Glucose is the brain’s primary energy source. However, before it enters the brain, it must cross a network of blood vessels called the blood-brain barrier. Glucose crosses the bloodbrain barrier by facilitated diffusion. In facilitated diffusion, a protein called GLUT-1 transports glucose across the endothelial cell membrane of the bloodbrain barrier to the brain. The expression of GLUT-1 can be regulated to alter glucose entry into the brain. When GLUT-1 is upregulated, the number of GLUT-1 proteins increases in the endothelial cells of the blood-brain barrier, increasing glucose entry into the brain. On the other hand, when GLUT1 is downregulated, the number of GLUT-1 proteins decreases, limiting glucose entry into the brain. Although the brain comprises only about 2% of
total body weight, studies have found that it consumes nearly 20% of the glucose in the body. Thus, understanding how the brain uses glucose is fundamental to understanding brain function. In a series of recent studies, Dr. Hwang and her lab investigated whether glucose entry into the brain is altered in patients with obesity and patients with Type 2 diabetes. One of the studies consisted of 25 participants: nine lean individuals, ten individuals with obesity, and six individuals with Type 2 diabetes. The participants with Type 2 diabetes received insulin intravenously to bring their glucose to baseline levels. Then, they received dextrose, a sugar similar in structure to glucose, to raise their blood sugar levels. Glucose levels were measured in a region of the brain called the occipital lobe [Figure 3]. Results showed that patients with obesity and Type 2 diabetes had lower glucose concentrations in the brain, possibly due to the downregulation of GLUT1.2 Dr. Hwang believes that reduced glucose entry into the brain in patients with diabetes and obesity could be a compensatory mechanism that protects the brain from high blood glucose levels, but long term could be associated with unwanted or detrimental consequences.
As a physician-scientist, Dr. Hwang draws inspiration for what scientific questions to ask from her patients. She believes that understanding how diabetes and obesity affect the brain
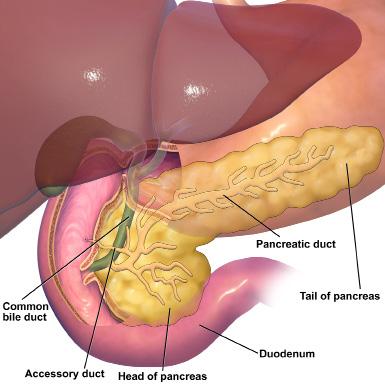

Figure 3. The occipital lobe is located at the back of the brain and is responsible for processing vision. Image courtesy of Wikimedia Commons.
will facilitate the development of new therapies to improve patient outcomes. By using cutting-edge brain imaging methods, she feels that she has a unique window into patient’s brains.
Obesity and diabetes continue to be on the rise due to many factors, such as lifestyle and genetics. However, by understanding their effects on the brain, their symptoms can be significantly improved, which Dr. Hwang strives to do. A fun but challenging aspect of conducting research studies is that Dr. Hwang finds she is always trying to develop new and better ways to answer a scientific question. It is really rewarding to be able to do a job where you can constantly learn new things.
1. “Global Obesity Rates Surpass One Billion, Analysis Finds.” News-Medical, 24 Feb 2024, https://www.news-medical.net/news/20240229/Global-obesity-rates-surpass-one-billion-analysis-finds.aspx#:~:text=The%20total%20 number%20of%20children,analysis%20 published%20in%20The%20Lancet.
2. Interview with Janice Hwang, MD, MHS, 2/2/2023.
3. Hwang, Janice J et al. “Blunted rise in brain glucose levels during hyperglycemia in adults with obesity and T2DM.” JCI insight vol. 2,20 e95913. 19 Oct. 2017, doi:10.1172/jci.insight.95913
Being faced with making a personal medical decision may seem daunting in the case of serious illness or injury, but having to make a sudden medical care decision for another person, such as a loved one, can be overwhelming. Dr. David Hwang, a neurologist working in the University of North Carolina at Chapel Hill’s neuroICU, is a prominent researcher in the field of neurology who studies neuroprognostication and the psychological impact these situations have on family members holding a decision-making role.
During his fourth year of medical school, Dr. Hwang completed his neurology rotation and fell in love with the specialty. He pursued a residency in neurology and found a passion for the intensive care unit (ICU) during his internal medicine internship. He enjoyed the fact that medical care in the ICU involved data driven decisions, coupled with the ability to help patients either overcome a significant illness or to pass away in a dignifying way. As he continued into the neurology portion of his residency, he decided to focus on a career in the neuroICU.
One of the most important parts of neurocritical care Dr. Hwang has conducted extensive research on is that of prognostication factors when treating patients with severe acute brain injuries. For patients with the most severe injuries in the neuro-ICU, families may need to decide on whether to pursue comfort care or prolong life care through a
tracheostomy (a breathing tube) and a feeding tube. These decisions often rely on the prognosis that clinicians determine and present to families when a patient is admitted to the ICU.
In neurocritical care, prognostic estimates can be generated using condition-specific calculators that use data regarding the state of the patient as well as demographics, such as age, to predict an outcome for the patient at various time points (such as threemonths, six-months, etc).
Dr. Hwang decided to investigate the accuracy of these calculators given the frequent use of them by clinicians.
“I think when most clinicians think about how to scientifically tell a family how a patient is going to do, the instinct is to try and use one of these calculators because it allows you to feel like you are not just guessing and only relying on your experience.”1
The calculator examined for the study is known as the ICH score. ICH

stands for intracerebral hemorrhage (bleeding in the brain), a kind of subacute brain injury.
The experiment involved asking doctors and nurses to write down their own three-month predictions for patients admitted to the neuro-ICU with an intracerebral hemorrhage, which was compared to the calculator three months later. Despite the fact that some clinicians may reference these calculators for assistance in prognosis, the results found that the calculator was less accurate in predicting patient outcomes than the clinicians who were directly involved in the patient’s care.
Dr. Hwang attributes the increased accuracy of the clinicians to the fact that there is more data available to the neurocritical care team, such as various monitors and conversations with family members, than the specific and uniform data points such as age and intracerebral hemorrhage that the calculator considers.2
“It’s reassuring on one level that there is some room from a clinician perspective to utilize your instinct about how you think patients are going to do and communicating that with the family. On the other hand, it might imply that we have a lot of room to make these calculators more accurate. Ideally, we wouldn’t want to say the best thing we have are just educated guesses.”1
Continuing the premise that clinicians have access to more factors than condition specific calculators, Dr. Dr. David Hwang

Hwang conducted a second experiment to better understand what specific factors clinicians considered. This consisted of asking doctors and nurses to write down the factors they were considering while going through the process of determining a prognosis.
The results showed that doctors and nurses approached prognostication differently from one another. The nurses relied more on the examination and observations of a patient off anesthesia, seeing if the patient was opening their eyes or moving around, as well as considering what the family told them about the patient’s character.
The doctors tended to focus on CT scans and radiographic imaging that showed the size and location of the stroke. Despite this difference in decision factors, the doctors and nurses were still equally accurate in predicting the outcome of the patient, indicating that there are multiple approaches to generating a prognosis.3
Along with prognostication calculators, Dr. Hwang has focused on how family members who are responsible for making a care decision approach decide on which form of care to pursue. Dr. Hwang and a group of researchers conducted a nationwide poll and asked individuals to imagine they had to decide the course of care for a parent who had sustained a brain bleed. They provided respondents with a list of factors that actual family members who had made these decisions considered and had the respondents rank them in importance of consideration to making their final choice.
Among the respondents, considerations for respecting their loved ones wishes and ensuring their family member was comfortable were near unanimous. However, a few subgroups formed based on their reported next most important consideration. These included: concern for an accurate prognosis, concern for agreement among
the surviving family members on the chosen care plan, and another group who had concern for the finances of care. The researchers expected prognosis to be a prominent consideration for individuals but did not expect other factors, such as finances, to be a primary concern. This research and the subsequent paper presented the idea that, as Dr. Hwang says, “when you’re talking to the families, the prognosis is [only] a piece of it. You may be talking about your calculator and what you think the prognosis is going to be, but that may not actually be what is driving the family’s decision.”1, 4
More recently, Dr. Hwang has been focusing on how families cope with having a relative in the neuro-ICU. In one study that was a collaboration between Yale—his former place of occupation— and The University of Massachusetts, they provided families with decision guide booklets that had detailed information regarding the options for prolonging care, such as more in-depth information of a tracheostomy and feeding tube, or the details of providing comfort care.
Unintentionally, the study found that those who were provided a decision aid were more likely to choose prolonged care measures. However, of the families whose loved one survived, they suffered from more stress, anxiety, and depression with a greater need for psychosocial support. What it also demonstrated according to Dr. Hwang was that, “it was less of a decision about what is right or wrong, and more about if you choose to do that, [prolong care through tracheostomy and feeding tube] there is a lot of opportunity for us as a medical field to support people even once they leave the ICU. People need ongoing support.”1, 5
Recently, Dr. Hwang has received a grant in collaboration with the psychology department at the Massachusetts General Hospital in Boston, Massachusetts to pilot a new program that provides
psychological support to family members after they have made the decision to prolong care. It will include online sessions with trained psychologists who will provide support as families navigate the psychological impacts of making these decisions for their loved ones.
Dr. Hwang is excited for the future of neuro-prognostication research and emphasizes that there are many emerging treatments in the field. He also expresses that, “if people are interested in the mind and brain, there has never been a better time, in my opinion, to go into neurology or psychiatry.”1 There are many more questions to be asked and researched in the field of neurology, and Dr. Hwang is playing a crucial role in improving the field of neurocritical care.
1. Interview with Dr. David Hwang 2/5/24
2. Hwang DY, Dell C, Sparks MJ, Watson T, Langefeld CD, Comeau ME, Rosand J, Thomas W.K. Battey, Koch S, Perez ML, et al. 2015. Clinician judgment vs formal scales for predicting intracerebral hemorrhage outcomes. Neurology. 86(2):126–133. doi:https:// doi.org/10.1212/wnl.0000000000002266.
3. Hwang DY, Chu SY, Dell CA, Sparks MJ, Watson TD, Langefeld CD, Comeau ME, Rosand J, Thomas W.K. Battey, Koch S, et al. 2017. Factors Considered by Clinicians when Prognosticating Intracerebral Hemorrhage Outcomes. Neurocritical Care. 27(3):316–325. doi:https://doi.org/10.1007/s12028-0170430-7.
4. Hwang DY, Knies AK, Mampre D, Kolenikov S, Schalk M, Hammer H, White DB, Holloway RG, Sheth KN, Fraenkel L. 2020. Concerns of surrogate decision makers for patients with acute brain injury: A US population survey. Neurology. 94(19):e2054–e2068. doi:https://doi.org/10.1212/ WNL.0000000000009406. [accessed 2023 Dec 19]. http://www.ncbi.nlm.nih. gov/pubmed/32341190.
5. Muehlschlegel S, Goostrey K, Flahive J, Zhang Q, Pach JJ, Hwang DY. 2022 Jul 19. A Pilot Randomized Clinical Trial of a Goals-of-Care Decision Aid for Surrogates of Severe Acute Brain Injury Patients. Neurology.:10.1212/ WNL.0000000000200937. doi:https:// doi.org/10.1212/wnl.0000000000200937.

Imagine a world where your beating heart could hold the key to understanding not only your individual health, but also the risks lurking beneath the surface of commonly prescribed medications. At the University of North Carolina at Chapel Hill, one Chapel Hill native has dedicated his research to just that. After completing his medical school education at UNC-Chapel Hill, Dr. Brian Jensen joined the School of Medicine faculty in 2009 and has been a pioneer in researching the alpha one adrenergic receptor.1 Through his projects, the field could reshape understanding of cardiovascular health and even save lives through alternative medical therapies.
Alpha one adrenergic receptors, or α1-ARs, are a type of receptor found on the surface of certain cells, including those in the heart and blood vessels.2 There are three subtypes of α1-ARs—the Jensen lab studies the α1A-AR subtype. These receptors are part of the sympathetic nervous system, which is responsible for the body’s fight or flight response. When activated by a neurotransmitter, either adrenaline (known as epinephrine) or noradrenaline (known as norepinephrine), alpha one adrenergic receptors trigger increased heart rate, constricted blood vessels, and heightened blood pressure.2 Within the heart specifically, alpha one adrenergic receptor regulate heart muscle metabolism and function to preserve the contractile function of the heart. Thus, understanding how these receptors function is crucial to unraveling the complexities of cardiovascular health and developing targeted therapies for heart-related conditions.
In his postdoctoral work, Dr. Jensen focused on catecholamines— monoamine neurotransmitters that are key to different stress responses. Due to his interest in the subject matter and the possibility of clinical applications, he continued
studying the effects of monoamine neurotransmitters on the cardiovascular system after arriving at UNC-Chapel Hill. Recently, Dr. Jensen was the head of a study involving Medicare patients who were taking alpha blockers (medications that block the effects of α1A-ARs) for prostate problems.1 Through this study, they discovered a significantly increased risk of cardiovascular diseases among male Medicare patients who took alpha blockers in the past year.1 This finding suggests potentially widespread risks of alpha blockers because they are often used to treat benign prostatic hyperplasia (roughly 5 million men in the U.S. take them). To mitigate these risks, Dr. Jensen believes that there may need to be “increased caution in prescribing these drugs,” and that risks need to be explored further.1
In order to further understand the roles α1A-ARs have in cardiovascular health, Dr. Jensen conducted a study involving different mouse models. Some of the mouse models were α1A-ARs KO mice, also known as “knockout,” mice. This means that the gene that codes for the α1AARs in the mice was removed, preventing the expression of the receptors. Dr. Jensen used two different groups of mice, one where the α1A-ARs were removed only in the heart, while the other group had the α1A-ARs removed throughout the whole body. Dr. Brian Jensen


In his work with these mouse models, Dr. Jensen and his team have found that lacking receptors all over the body significantly decreases the heart’s oxygen consumption. Jensen also looked at mitochondrial function using a technique known as advanced respirometry. Advanced respirometry measures how much oxygen a cell is consuming to assess cell function. Low oxygen consumption suggests issues with cell’s metabolism. Overall, Dr. Jensen observed decreased oxygen consumption in the global KO mice and increased mitochondrial oxygen function in normal mice with α1A-ARs receptors when the receptors were activated with an agonist.2
Dr. Jensen was surprised by some global KO mice findings. Previously, it was thought that a difference in oxygen consumption would only be detectable while the mice were undergoing a stressful event. However, his results show that the decreased function was consistent, regardless of the presence of a stressor. Additionally, the KO that lacked the α1A-AR only in heart muscle cells had dramatically negative effects resulting from the initiated cardiac failure—most of these mice died within 5 days of the cardiac episode.1 These findings parallel the Medicare study, which found an increased cardiac risk after taking alpha blockers.3
With his findings, Dr. Jensen believes that developing and testing new agonist drugs that activate α1A-ARs may be valuable route to increasing the effectiveness of heart failure treatments.1 Using the information about alpha 1 receptors having adverse metabolic profiles, Jensen is now focusing on the therapeutic potential of an α1A-AR agonist, which could have applications in the treatment of cardiovascular diseases. Overall, Dr. Jensen is hoping to catalyze a shift towards treatment-oriented research,
using his lab’s work to identify potential new targets. He also emphasizes the importance of a multidisciplinary approach to this research. As Jensen said, “I wouldn’t have been able to do this without the collaboration of a lot of really talented people at UNC.” His pioneering research in cardiac pharmacology not only sheds light on the intricate molecular mechanisms underlying cardiovascular health, but also may pave the way for transformative therapies that have the potential to change the landscape of cardiac medicine.
1. Interview with Dr. Brian Jensen, MD 2/19/24.
2. Zhang, J., Latour, C.D., Olawore, O., Pate, V., Friedlander, D.F., Stürmer, T., Funk, M.J. and Jensen, B.C., 2023. Cardiovascular Outcomes of α-Blockers vs 5-α Reductase Inhibitors for Benign Prostatic Hyperplasia. JAMA Network Open, 6(11), pp.e2343299-e2343299.
Imagine having to make over 20,000 decisions in your day, and each decision you make affects your appearance, personality, and health. While this may seem to be an overwhelming and impossible task, the 20,000 genes coded by DNA in your cells do just this process to express all the traits that make up who you are. The long and winding double-helix most people are likely familiar with as DNA, houses the human genome. Our genome contains genes, which determine the traits of our body that appear as physical characteristics or even our personality and behaviors. Out of the 20,000 genes in the human genome, some of the most important are the pieces of DNA that code, or create the instructions for, bodily proteins. Proteins serve multiple functions in the body, such as metabolizing food, aiding in cell processes, and growing and repairing body tissues. Occasionally, a defect or abnormality may occur if the coding sequence for the protein becomes mutated or if a coding error occurs. Genetic fetal abnormalities, which are often difficult to find the cause of, occur as soon as the DNA is formed during conception. Though abnormalities can physically appear as soon as the fetus develops, the cause of them at the genetic level is often difficult to detect.
Birth defects due to genetic abnormalities affect 3 to 4 percent of all pregnancies and often change the way the pregnancy is monitored and the process of delivery.1 Luckily, due to recent technological advances, we can detect unexplained fetal abnormalities in the prenatal stage by determining if there is a genetic error in the genome of the fetus. By being able to diagnose fetal abnormalities in the prenatal stage, maternalfetal health counseling can be
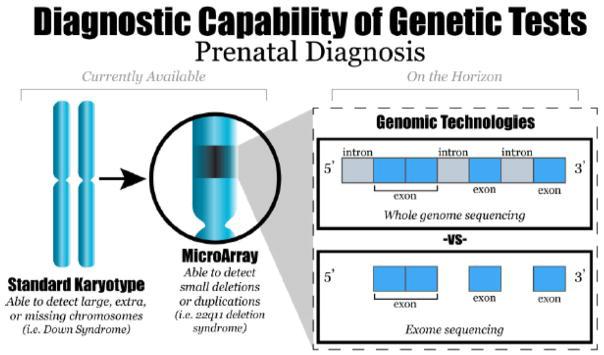
1. A diagram of previously available karyotype genetic testing versus recent exome-sequencing technology. Image courtesy of Hardisty EE and Vora NL.

improved. Scientists, using a new type of genome sequencing technology, can determine if there is a genetic error in the genome of the fetus. “We as humans have about 20,000 genes, and we only understand what about maybe 6,000 of those genes do,” explains Dr. Neeta Vora, “…so I became really interested in trying to understand what’s going on with the fetus [and] what’s going on with human development.”1 Dr. Vora is a Professor of MaternalFetal Medicine and Genetics and the Director of Reproductive Genetics at The University of North Carolina at Chapel Hill. She completed her medical residency at the Tufts Medical Center in Boston, Massachusetts, and now is board-certified in Obstetrics and Gynecology, Maternal Fetal Medicine, and Clinical Genetics. Dr. Vora sees patients that have pregnancies where the fetus has a birth defect identified by an ultrasound. She obtains DNA from the fetus through different types of diagnostic testing. For example, amniocentesis, or when a sample of amniotic fluid is extracted from the womb, can be used for genetic testing, specifically sequencing techniques. Genome sequencing examines the order of base pairs that make up DNA.
Right here at UNC, there is a DNA sequencing laboratory capable of a newer type of sequencing technology called
exome sequencing, a process that identifies variants in the DNA that cause birth defects. The genome contains introns and exons. Exons are the protein coding regions of the genome and account for only about 1.5 percent of the whole genome. Introns are pieces of noncoding DNA found between exons. Exome sequencing is the analysis of DNA base pairs in only the protein-coding exons of the genome (Figure 1). By conducting sequencing for the exons, Dr. Vora and her team are able to locate novel genes of interest that may contribute to fetal development and figure out which exact gene, if there is a mutation present, causes an abnormality in certain patients. Although exome sequencing is more costly and has a longer turnaround time, it has a higher diagnostic yield since it can reveal a more detailed sequence at a higher resolution. The patients Dr. Vora sees are then more likely to receive an explanation for the fetal abnormality if there is a genetic cause.
Exome sequencing may also reveal the purpose of unknown genes, including identifying genes critical to human development. After exome sequencing, bioinformatics filters are used to identify pathogenic variants to see if the fetal brain abnormality is caused by a genetic error1 (Figure 2). Exome sequencing of fetuses with birth defects may also reveal novel genes critical to human development. Standard genetic testing with a karyotype or microarray has a much lower resolution than sequencing (Figure 1). This is because a karyotype only shows deletions (missing parts) on a chromosome or if there is an extra copy of a chromosome. A microarray can be used to show levels of expression of known genes, but novel genes may be difficult to show in a microarray if the role they play in development has not yet been discovered.
Dr. Vora, along with a large team of researchers around the country, use exome sequencing technology that looks specifically at protein-coding regions of the DNA, called exons. They have used zebrafish to edit or silence, which is done by turning off certain genes, genes of interest to human development to identify whether the zebrafish has the same abnormalities seen in the fetus. Her lab focuses specifically on fetal brain anomalies. “[Zebrafish] are very experimentally tractable, you can easily [manipulate] them, silence genes, [and] you can use gene editing technologies,” remarks Dr. Vora, “…and we share 70 percent of our genes…with the zebrafish.”1 Zebrafish (Danio rerio) are a small translucent fish and have embryos that are easy to examine. To observe whether a gene causes a birth defect, genes of interest are either silenced or edited in
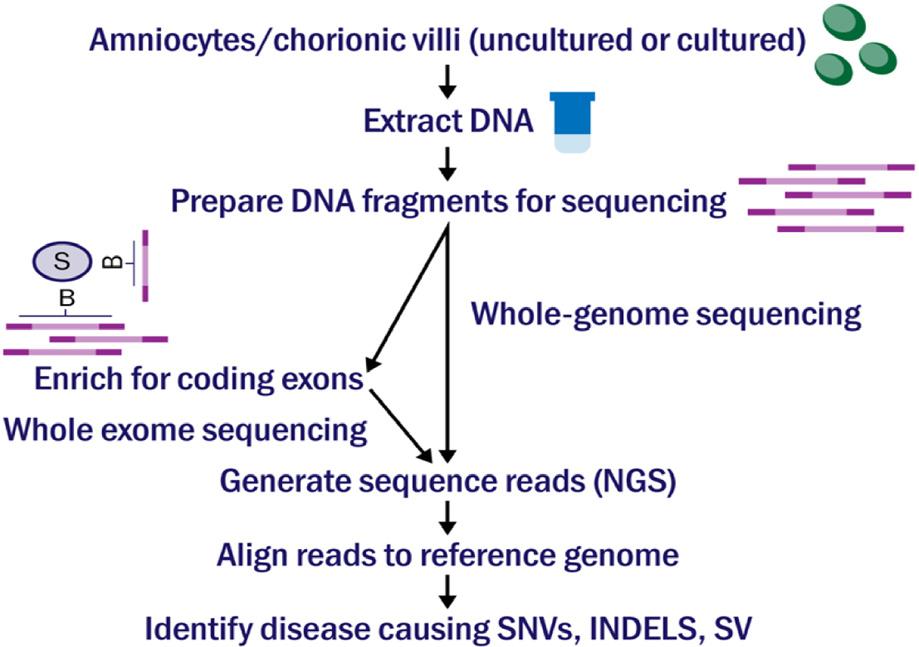
2.
exome sequencing to identify a disease. Image courtesy of Goh and Choi.

Figure 3. Photo of zebrafish modeling, including images and charts showing the effect of a variant MAPK4 protein on fetal development. Figure courtesy of Patterson et al.
zebrafish embryos and observed after about 3 days for physical abnormalities.3 If the zebrafish have abnormalities in brain development like the fetus, this provides evidence that the gene is important for human brain development.1 Using this new exome editing technology, Dr. Vora and her team have already identified several new genes that affect normal fetal brain development. For example, a gene that codes for a protein called MAPK4 has been found to cause zebrafish heart and craniofacial defects in its embryo stage when the gene is silenced (Figure 3).3 As a physician scientist, Dr. Vora’s research is driven by her patients1. Her progress on exome sequencing in the prenatal stage of fetal development has led to new answers and potential closure for her patients. Knowing what is causing the fetal abnormality helps improve prenatal counseling and enables the pediatrician to prepare for delivery.4 Dr. Vora explains that “The most exciting thing is when we discover a novel gene that was previously unknown, and then we publish on [the novel gene] and we can go back to the family and say, ‘We found the answer for why this happened in that pregnancy.’”1 Using prenatal sequencing, our ability to understand the genetic causes of congenital anomalies has increased tremendously.
1. Interview with Neeta Vora, M.D. 02/05/24.
2. Vora, N.; Norton, M. E. Prenatal exome and genome sequencing for fetal structural abnormalities. Am. J. Obstet. Gynecol, 2023, 228(2), 140–149.
3. Patterson V.; Ullah F.; Bryant L.; Griffin JN.; Sidhu A.; Saliganan S.; Blaile M.; Saenz MS.; Smith R.; Ellingwood S.; et al. Abrogation of MAP4K4 protein function causes congenital anomalies in humans and zebrafish. Sci Adv, 2023, 9(17), eade0631.
4. Jelin, A.C.; Vora, N. Whole Exome Sequencing: Applications in Prenatal Genetics. Obstet Gynecol, 2018, 45(1), 69-81.

While it may seem obscure, answering the question of why people die of cancer is surprisingly quite plain. Cancer spreads uncontrollably and we often cannot stop it. However, the solution to such a straightforward pathology has proven to be one of the most elusive problems in medical research. Many current treatments focus on trying to shrink cancer tumors. These existing therapies can serve that purpose, but they usually do not completely eradicate these cancerous masses. Consequently, the tumors eventually become resistant to those treatments, and then patients have to receive a different therapy. Eventually, each treatment that the patient moves on to is likely to become less and less effective. This allows the tumors to continue growing and then they can spread and take over the body. Wherever these cancer cells are, those functions shut down. Regardless if the cancer is in the brain, bones, liver, or anywhere else, it just completely takes over.
During his sophomore year of college, Dr. Chad Pecot contracted testicular cancer.

Initially, by just cutting out the cancerous tumor, it seemed like surgery would fix everything. However, after receiving a scan about
a month after the surgery, Dr. Pecot was alerted that the cancer had spread to his abdomen. This changed everything because at first, the mission was just to cut out the cancer and move on. Something that seemed like a simple surgery, led to months of hospital visits to receive blood work and chemotherapy. Being thrown into having it and then being surrounded by those who were also experiencing cancer was Dr. Pecot’s inspiration to pursue a career in oncological research.
Dr. Pecot’s current research consists of studying how cancer spreads and what therapies can be created to stop that. In the last 15 to 20 years, most cancer research has been focused on only around 2% of our genome. When looking at what gets transcribed from the DNA, only about 2% translates into protein. Therefore, the pharmaceutical industry has been very focused on making drugs that target these proteins and producing many effective drugs with
this focus. There are many new medicines now that usually block the function of a particular protein. Therefore, oncogenic – or cancerous – proteins that grow and spread, can be blocked by a therapy that blocks the protein expression. This has been an effective therapeutic for cancer, with the last 15 years of progress in cancer research focusing on this avenue. The irony is that only 2% of what becomes transcribed is becoming protein.
Dr. Pecot’s lab has ventured into studying the other 98% of the genome. These are transcribed into non-coding RNAs, such as miRNAs, circle RNAs, and snoRNAs, which do not code for proteins. There are many different types of RNA that the tumors are taking advantage of to grow and spread, or metastasize; this is a major area of interest in the Pecot Lab. Therefore, they hope to bridge a gap in cancer treatment, as there are no drugs to mitigate metastasis or tumor involvement with RNA. Still, there is a lot of promise in
these non-coding RNAs, both in trying to deliver or silence them. The Pecot Lab is interested in creating RNA medicines to target metastasis through various interference projects. Dr. Pecot describes these treatments as, “Medicines made out of RNA to target RNA.” The Pecot Lab conducts projects that develop novel therapeutics out of RNA which are engineered to go into the tumors and target the RNA that are facilitating metastasis through key receptors. Delivering RNA to cancer is not a new idea because researchers have known that RNA could be capable of targeting challenging problems in tumors. There has been a lot of research done on the specific genes that contribute to cancer, which are challenging to target. This means that many in the pharmaceutical industry have unsuccessfully tried to make drugs that hit the protein directly. Even though it is well known that the protein needs to be eradicated, it is extremely difficult. So, while monoclonal antibodies and small molecule inhibitors have been a profound step forward in oncology treatments, many targets in cancer are regarded as “undruggable” using these approaches. These targets could include transcription factors, difficult-to-target proteins (like mutant KRAS or MYC), or specifically non-coding RNAs. However, The Pecot Lab is exploring potential ways for RNA to tackle these targets.
The Pecot Lab is working to solve the problem of delivering the RNA therapeutic. By engineering RNA
and putting certain modifications on molecules, the Pecot Lab is strategizing avenues for the RNA to eradicate the tumor and not cause a negative immune reaction. The body will potentially mistake the RNA medicine for a virus and have a negative reaction to it. The focus
“[Dr. Pecot’s Lab] hope to bridge a gap in cancer treatment, as there are no drugs to mitigate metasis or tumor involvement with RNA”
of the lab’s work is to engineer an RNA therapeutic that prevents the patient’s immune system from having a large response. The lab also aims to make the RNA much more stable because unlike DNA, which is very stable, RNA degrades quickly. The majority of RNA engineering goes into both keeping the molecule in “stealth” mode (avoiding a negative immune reaction) and also, to keep it from degrading very quickly to enable the medication to work.
Previously, many people in the cancer research community did not fully support RNA use in these matters. However, because of the pandemic, there has been growing interest surrounding the idea that there are technologies that

can make effective RNA medicines. “Many people have received mRNA vaccines and that is only one of the different types of RNA medicines that people have been working on for a long time, myself included, and hopefully it has now made the world realize RNA can be an effective medicine in very little time,” Dr. Pecot says. “I think that what we’re working on is very similar. These could be effective medicines against cancer as well.”
Dr. Pecot believes the future of RNA research in cancer lies in improving delivery methods. The Pecot Lab is pioneering methods to reach the “undruggable” cancer targets not only through physical testing but also through artificial intelligence. The intersection of artificial intelligence and RNA medicine design to create accurate sequences and structures for inducing the desired effects is a reflection of the exciting direction the cancer research field is moving. Similarly, the promise in protein engineering to attach RNA sequences directly will also transform cancer treatment because this allows the protein to guide the RNA and bind the receptors. A concept that could eradicate cancerous tumors faster. This is a point of interest for Dr. Pecot’s further cancer research, continuing to formulate clever strategies to deliver RNA therapies and hit stubborn targets. The potential success of these RNA-based therapeutics can significantly impact the future of cancer treatment and revolutionize patient care through additional therapy modalities.
1. Interview with Chad Pecot, MD. 2/27/2024.
2. Bowler, M. M., Glavatskikh, M., Pecot, C. V., Kireev, D., & Bowers, A. A. (2023). Enzymatic Macrolactamization of mRNA Display Libraries for Inhibitor Selection. ACS Chemical Biology, 18(1), 166–175.
3. Papke, B., Azam, S. H., Feng, A. Y., Gutierrez-Ford, C., Huggins, H., Pallan, P. S., Van Swearingen, A. E. D., Egli, M., Cox, A. D., Der, C. J., & Pecot, C. V. (2021). Silencing of Oncogenic KRAS by Mutant-Selective Small Interfering RNA. ACS pharmacology & translational science, 4(2), 703–712.

Within the vast field of nutrition research, a groundbreaking approach is gaining momentum: precision nutrition. Defined by its focus on tailoring dietary recommendations to individuals rather than broad populations, precision nutrition holds promise for revolutionizing how we approach health and wellness. Precision nutrition, akin to precision medicine, focuses on individualized dietary recommendations that are linked to specific genetic, behavioral, and environmental factors.
Dr. Susan Sumner, a Professor of Nutrition at UNC Chapel Hill’s Nutrition Research Institute (NRI), and the Director of the Metabolomics and Exposome Laboratory (MEL) at the Univesrity of North Carolina at Chapel Hill, sheds

light on the significance of precision nutrition and its potential impact on diverse communities. Drawing insights from genomics, metabolomics, and behavioral sciences, Dr. Sumner seeks a multidisciplinary approach in exploring the connection between heredity, lifestyle factors, and dietary interventions.
The Metabolomics and Exposome Laboratory (MEL) at the Nutrition Research Institute (NRI) is at the forefront of utilizing untargeted metabolomics/exposome approaches to explore pharmacological and nutritional targets for the development of strategies. Metabolomics is the study of small molecules, called metabolites, in biological samples such as cells, tissues, or fluids. Metabolites are produced by the body’s metabolism, and can provide insight into our processes and disease status/capability. Exposome approaches involve studying the totality of environmental exposures that an individual experiences throughout their lifetime, including factors like diet, lifestyle, medications, pollutants, and stressors. This strategy aims to understand how these exposures influence health and disease outcomes.
The MEL collects extracts from cells, tissues, or biological fluids from in vivo or in vitro studies, subjecting them to

untargeted metabolomics analysis. This data is then analyzed using phenotypic anchors obtained from tissue culture studies or in vivo models, including factors such as health status, dietary intake, medications, and genetic variants,

among others. This approach allows Dr. Sumner and the other researchers in the MEL to explore the role of metabolism in their studies and then apply this to nutrition.
Precision nutrition incorporates multi-omics methods that delve into various layers of biological information, including the genome, proteome, metabolome, and microbiome. This provides individualized insights into the mechanisms behind dietary strategies and further fosters a deeper understanding of metabolism at the chemical level.
"Precision nutrition focuses on the individual, tailoring dietary needs to specific genetic, behavioral, and environmental factors," explains Dr. Sumner. Similar to precision medicine, which customizes medical treatments based on individual genetic makeup, precision nutrition aims to provide personalized dietary prescriptions to optimize health outcomes. “It is about honing in on the unique needs of each individual, rather than adopting a onesize-fits-all approach,” says Dr. Sumner.1
Dr. Sumner underscores the importance of precision nutrition in tackling health inequalities and enhancing overall well-being within
various communities. She emphasizes that by taking into account elements like genetic tendencies, environmental factors, and cultural aspects, precision nutrition provides valuable insights that can guide particular approaches to enhance health results among several different population groups.
Yet, navigating the complexities of precision nutrition research poses challenges. One of the key pieces in precision nutrition research lies in deciphering the complex interactions between genetics, metabolism, and dietary intake. "Integrating data from various sources, including genomic sequencing, metabolomics profiling, and dietary assessments, is essential to develop comprehensive profiles of individual nutritional needs," she adds.
Furthermore, the implementation of precision nutrition approaches holds promise for transforming clinical practices. Dr. Sumner stresses the importance of integrating personalized dietary advice into regular healthcare evaluations, enabling patients to make educated decisions regarding their eating habits and overall health. Her research underscores the importance of collaboration across disciplines in advancing precision nutrition initiatives, by bridging the gaps between nutrition, genetics, and clinical practices.

Looking ahead, the integration of metabotyping into clinical practice represents a significant advancement. However, challenges such as data integration and implementation remain. Collaborative efforts and continued research are essential to realize the full
potential of precision nutrition. Diet and disease are intricately linked, and precision nutrition represents a paradigm shift in how we approach dietary recommendations and health promotion. By harnessing the power of personalized dietary prescriptions, precision nutrition has the potential to revolutionize healthcare, improve health outcomes, and empower individuals to take control of their nutritional wellbeing. Precision nutrition is paving the way for a healthier future.
References
1. Interview with Susan Sumner, Ph.D. 1/27/2024
2.Woychik, R. (2023, February).
Precision nutrition improves health at individual level, expert says. Environmental Factor. Retrieved from https://factor.niehs.nih.gov/2023/2/ feature/4-feature-precision-nutrition

Human immunodeficiency virus (HIV) is an infection that attacks the body’s immune system, our body’s natural defense, ultimately reducing its efficacy in fighting back diseases. The HIV epidemic in Sub-Saharan Africa (SSA) continues to be one of the largest ongoing epidemics in the world today, with SSA patients accounting for two-thirds of people living with HIV globally.1 HIV is associated with a variety of disorders, including cancer and lymphoma. Lymphoma refers to cancers found in the lymphatic system, which account for a significant portion of cancers in this context. Unfortunately, in SSA where the burden of HIV is high, critical information on the intrinsic and extrinsic pressures under which HIV-associated lymphoma arises is lacking (Figure 1).
In 1995, Antiretroviral Therapy (ART), which consists of a combination of drugs that repress HIV replication, was first introduced to treat HIV patients.2 Since then, with more HIV patients worldwide having access to ART, there has been an increase in the HIV survival rate. Despite an increasing worldwide population of ART-experienced HIV lymphoma patients, limited
information is known on the different characteristics under which these patients’ tumors arise compared to those of HIVnegative patients or ART-inexperienced HIV patients (Figure 2). Over the course of his career and working in collaboration with partners in Malawi, Dr. Yuri Fedoriw, Co-Director of the University of North Carolina at Chapel Hill Project MalawiCancer Program, was exposed to the lack of information known on HIV-associated lymphoma for both ART-experienced and ART-inexperienced patients. Hence, these medical issues that needed to be urgently addressed on a global scale inspired Dr. Fedoriw to establish the research laboratory where students, clinicians, and researchers at UNC-Chapel Hill and Kamuzu Central Hospital in Lilongwe, Malawi work to enhance the current understanding of HIV-associated lymphoma. He stated, “Our research is particularly valuable in a country of need and the questions we ask are relevant to the local context. In SSA, HIV is a generalized epidemic affecting all demographics, in contrast to the United States where HIV is largely concentrated in marginalized communities that we cannot yet effectively reach through healthcare. So, if you are trying to understand what will improve care for this (Sub-Saharan African) population, it is not always possible to draw parallels to the US context.” Therefore, due to the remarkably different conditions under which the disease arises in the two continents, Dr. Fedoriw emphasizes that generalizing treatment methods can be detrimental to care. Dr. Fedoriw’s lab has conducted many studies, but one of the noteworthy studies conducted is the identification of prognostic biomarkers and transcriptional profiles of HIV-associated diffuse large B-cell lymphoma (DLBCL). Identifying prognostic biomarkers is essential to give an indication of the likelihood of disease progression, and transcriptional biomarkers allow quantification of gene expression in tumors. Non-Hodgkin lymphoma is one of the most frequent HIV-associated lymphomas, and DLBCL is its most common subtype.4 In this study, whole transcriptome sequencing (which allows for a

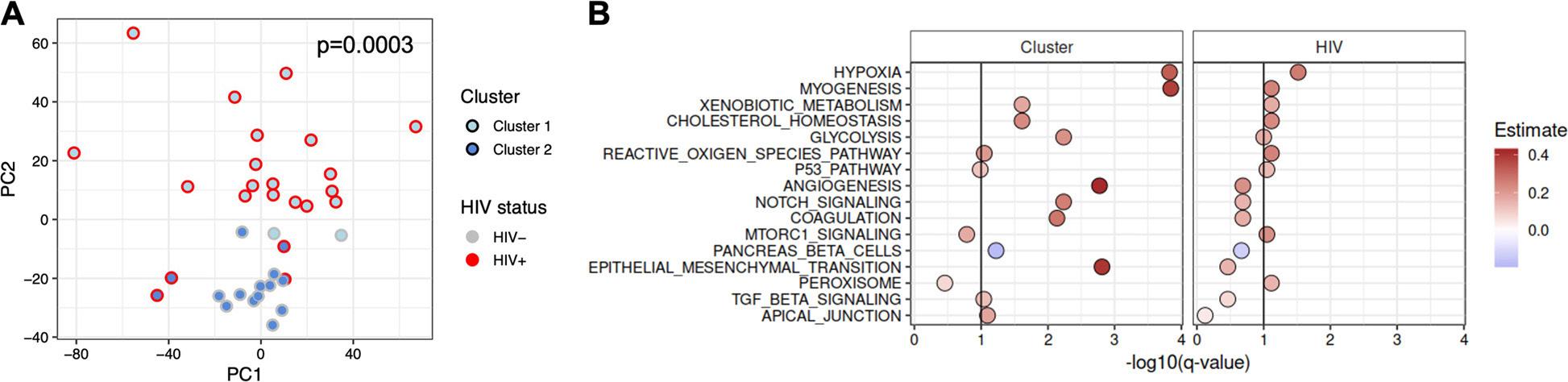
Figure 3. (A) RNA sequencing data demonstrating two clusters with differential gene expression (light blue and dark blue). (B) When overlaid, patients’ HIV status (red and gray) strongly correlates with cluster. Image courtesy of Fedoriw Lab.
complete picture of RNA transcripts) was completed to see the differences in gene expression between HIV-positive and HIVnegative DLBCL cases. The results showed that HIV-associated DLBCL was enriched for hypoxia (state where not enough oxygen is available for tissues) induced genes, angiogenesis (formation of new blood vessels) induced genes, and expression modules related to oxidative stress. Tumor hypoxia and angiogenesis develop due to increased cell proliferation and abnormal tumor blood vessels leading to reduced oxygen and nutrients.5 The results also showcased significant differences related to oxidative stress and these findings exemplify the biological differences in HIV-associated DLBCL. Moreover, surprisingly three out of four HIV-positive DLBCL cases that clustered with the HIV-negative cases had undergone relatively long durations of ART before lymphoma diagnosis. The findings suggest that the tumors of ART-experienced HIV patients may have more in common with HIV-negative tumors than the tumors of ART-inexperienced HIV patients (Figure 3).
Regarding prognostic signatures, in HIV-positive DLBCL cases of the cohort, it was found that higher interferon (IFNs) response is associated with better DLBCL outcomes.6 IFNs are proteins that belong to cytokines, which are a group of signaling molecules involved in the upregulation of the immune response.7 Additionally, overexpression of both BCL2 and MYC proteins (proteins involved in the regulation of cell proliferation and cell death) is associated with reduced survival regardless of HIV status.8 These results affirm the need for prognostic markers specific to HIV status and global regions to derive clinical conclusions.6
With most HIV-associated lymphoma research in the world being done on HIV-negative patients, there is a demand for understanding the differences in the immunologic and genetic mechanisms under which HIV-associated DLBCL arises, not only in SSA but also worldwide. Additionally, with the HIV survival rate increasing due to ART and ART-experienced HIV individuals becoming the predominant subpopulation of all individuals with HIV, there is also an increased demand for understanding the post-treatment effects that ART has on the tumor development of HIV-associated DLBCL. Dr. Fedoriw aims to bridge this gap with his research. With so many groundbreaking analyses being done by clinicians, researchers, and students in the lab, Dr. Fedoriw says they are undoubtedly the best part of the lab and that “it is awesome to have such creative and intelligent conversations with motivated people.” Dr. Fedoriw hopes that in the future, the lab’s findings may lead
to novel therapies for people living with HIV and lymphoma worldwide (Figure 4).

Figure 4. UNC Project Malawi Program Building and Members. This new structure is the home to program leadership and the pathology laboratory. Image courtesy of Fedoriw lab.
1. Moyo E, Moyo P, Murewanhema G, Mhango M, Chitungo I, Dzinamarira T. Key populations and Sub-Saharan Africa’s HIV response. Front Public Health. 2023, 11, 01.
2. Antiretroviral Therapy - PAHO/WHO | Pan American Health Organization. https://www.paho.org/en/topics/ antiretroviral-therapy (accessed February 23, 2024).
3. Interview with Dr. Yuri Fedoriw, M.D. 02/21/2024
4. Muz B, Puente P, Azab F, Azab AKw. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Dove Press. 2015, 3, 83-92.
5.Huguet M, Navarro JT, Moltó J, Ribera JM, Tapia G. Diffuse Large B-Cell Lymphoma in the HIV Setting. Cancers 2023, 15, 3191.
6. Identifying transcriptional profiles and evaluating prognostic biomarkers of HIV-associated diffuse large B-cell lymphoma from Malawi. Modern Pathology. 2020, 33, 14821491.
7. Khanna NR, Gerriets V. Interferon. In: StatPearls. StatPearls Publishing; 2024. Accessed February 23, 2024.
8. Riedell PA, Smith SM. Double hit and double expressors in lymphoma: Definition and treatment. Cancer 2018, 124, 4622-4632.

Perhaps you have heard that there are more microbial cells squirming about your body than human cells. Maybe you have been taking daily prebiotics in light of all the news about the gut microbiome. But how much do you know about the vaginal microbiome? Although the vagina’s microbiome rarely makes headlines, this diverse, complex ecosystem plays a crucial role in pathogen protection and genital health. Consequently, disruptions in the vaginal microbiome have grave implications, including increased susceptibility to preterm birth for pregnant individuals and sexually transmitted infections, such as HIV.1
Despite its vital role, the vaginal microbiome has been under-researched and treatments for its disruptions are outdated and relatively ineffective.2 However, Dr. Indriati Hood-Pishchany, a new physician faculty member of the University of North Carolina at Chapel Hill’s Pediatric Division of Infectious Diseases, aims to investigate the biome’s complexities and provide insight into treatments. Dr. Hood-Pishchany first developed a passion for research and discovery as an undergraduate at Houghton College.3 Later, while completing her M.D. and Ph.D. in Pathology, Microbiology, and Immunology at Vanderbilt University,3 her interest in infectious disease and its clinical conundrums grew4. Her specific interest in the vaginal microbiome emerged during her residency and fellowship after working with teen parents and learning about the microbiome’s role in pregnancy complications, such as preterm birth.4 With her microbiology background and her passion for addressing women’s health, infectious disease, and preterm birth, Dr. Hood-Pishchany works to understand the vagina’s microbes and develop pre-clinical models that address associated diseases.
The Hood-Pishchany lab has two main branches: 1) investigating the fundamental microbiology of the vaginal ecosystem and 2) using a bioreactor Dr. Hood-Pishchany adapted to grow vaginal communities.2 In the bioreactor, nutrient-rich media pumps in to support bacterial cultivation while waste is removed, a process closer to the steady-state system that human bacteria experience.4 A measurement called optical density—which measures the amount of light absorbed by a solution—monitors bacterial growth.4 Replicating the stable environment of humans is key to understanding microbe

metabolism and interactions, which ultimately informs the development of therapies to treat infections. For instance, if a researcher wanted to test a new antibiotic to treat bacterial vaginosis (BV), a condition affecting over 20% of women that can increase the risk of STIs and preterm birth,1 simply growing bacteria in the presence of antibiotics would not provide an accurate picture. Outside a steady state, bacteria growth rates differ, and interactions are altered. The bioreactor, however, helps mimic the dynamic reactions of communities exposed to antibiotics and/or probiotics in humans.
The Hood-Pishchany lab is currently working to develop the bioreactor model and validate its accuracy in predicting microbial dynamics and response to therapy in humans. The lab collaborates with the Vaginal Microbiome Research Consortium, a group of investigators working to expand knowledge of the vaginal microbiome and develop targeted therapies.4 The lab will assess the bioreactor in parallel with an ongoing clinical trial, called VIBRANT, which is testing a vaginally inserted tablet containing Lactobacillus crispatus (a “helpful” bacteria with antimicrobial properties1) to treat BV.5 Dr. Hood-Pishchany’s lab will test microbial communities representative of those observed in trial participants and compare how the therapy works in vivo versus in the bioreactor.2 If the bioreactor successfully replicates the microbes’ interaction with the therapy as seen in humans,
the implications are exciting. Dr. Hood-Pishchany explains that the bioreactor could help tweak the therapy’s components or aid researchers in targeting bacteria particularly resistant to existing therapies.4 The bioreactor does not replace human trials but rather provides a “scalable platform”4 to explore therapeutics. If a molecule that has never been clinically studied in humans demonstrates promising results in the bioreactor, researchers would prioritize and clinically explore it, overall streamlining the process of therapeutic development.
While still in the model-development phase, the bioreactor has already shown great promise. It can grow communities that imitate important features of the human vaginal microbiome.2 Working with so many microbes, however, is challenging and nuanced. First, it is difficult to determine which strains of bacteria best model in vivo communities. Second, Dr. Hood-Pishchany notes that using different species further complicates the work, explaining that “[As] you work with not just many strains but many species, they all have very different growth characteristics and different properties, and trying to standardize all that to make a reproducible complex microbial community is challenging.”2
1. Example of the bioreactor assembly for culturing vaginal communities. Figure provided by Dr. HoodPishchany.
However, this complexity makes the bioreactor such an exciting model. Previous work has taken more reductionist approaches to understanding diseases by, for instance, only focusing on one organism, such as Gardnerella (the bacteria predominantly seen in BV).2 However, BV is caused by whole communities and is affected by Gardnerella’s interaction with other community members.2 Thus, Dr. Hood-Pishchany’s encompassing approach provides critical insight into a field that, unfortunately, sees little innovation. Dr. Hood-Pishchany notes that the standard of care for BV has not changed since the 1980s, and treatments are not particularly effective as there are high rates of recurrence.2 She suggests the lack of innovation is partially attributed to BV primarily affecting women and
because it is construed as simply a nuisance—despite its high prevalence and association with negative health outcomes.1 Thankfully, Dr. Hood-Pishchany and other researchers are on the case. She remarks that it is “an exciting time to be in the field.”2
2. Top image shows normal vaginal bacteria; bottom shows bacteria seen in BV. Image courtesy of Wikimedia Commons.
In the future, Dr. Hood-Pishchany also hopes to explore how the menstrual cycle shifts bacterial composition. Research shows that during menstruation Lactobacillus jensenii and Lactobacillus crispatus decrease while Gardnerella vaginalis increases.6 Dr. Hood-Pishchany wants to study such shifts and aspires to collaborate with material scientists to create menstrual products that promote the retention and recovery of Lactobacillus and/or prevent harmful communities. Indeed, further innovations are in the lab’s future, and it will be exciting to watch its development. The team’s work to understand the microbiome’s complexities and accelerate therapeutic development will have significant impacts on women’s health. In a biome historically ignored and in critical need of advancement, the lab is here to make breakthroughs.
1. Chen, X.; Lu, Y.; Chen, T.; Li, R.; Front Cell Infect Microbiol. 2021, 11, 1.
2. Interview with Dr. Hood-Pishchany, MD, PhD. 02/07/2024.
3. UNC School of Medicine: Pediatrics. Welcome New Physician Faculty: Indriati Hood Pishchany, MD, PhD. https://www.med.unc.edu/pediatrics/2023/07/indriati-hoodpishchany/ (Accessed February 20th, 2024).
4. Correspondence with Dr. Hood-Pishchany, MD, PhD. 02/22/2024.
5. Mass General Brigham. Participants Needed! Vaginal Probiotic Clinical Trial to Prevent Recurrent Bacterial Vaginosis. https://rally.massgeneralbrigham.org/study/ vibrant (Accessed February 17th, 2024).
6. Srinivasan, S.; Liu, C.; Mitchell, C.; Fiedler, T.; Thomas, K.; Agnew, K.; Marrazzo, J.; Fredricks, D.; PLoS One 2010, 5, 1.
Innovation and scientific exploration can exist anywhere, including within infants, where you can find the focus of groundbreaking research with the potential to save preterm babies. The Good Lab at the University of North Carolina at Chapel Hill studies necrotizing enterocolitis (NEC), an intestinal disease predominantly affecting premature infants.1 The disease can cause intestinal epithelial cell damage, inflammation, multisystem organ failure, and even death. However, a lack of a complete understanding of the physical expression of the disease has prevented advances in the treatment NEC for the past 50 years.2
The head of the Good Lab and the Chief of the Division of Neonatology at UNC Children’s Hospital, Dr. Misty
 Dr. Misty Good
Dr. Misty Good

Good, was first exposed to NEC during her pediatric residency. She recalls the devastation of witnessing a preterm infant she was caring for unexpectedly pass away overnight, despite having appeared relatively healthy with no indication of this deadly outcome. She explains, “When that keeps happening in your clinical world, and you’re interested in science-…-you really want to make sure that you figure out what could we be doing differently.”2 Dr. Good transformed the loss she experienced in the clinical world into motivation for her research as a neonatology fellow at the University of Pittsburg–and beyond. She became invested in battling NEC and joined a laboratory studying the disease as a fellow. Dr. Good began her journey into the field of basic research by learning to pipette water, but quickly advanced to exploring complex models and discovering new techniques to pursue her own curiosities. For example, she created an innovative method using a human breast pump to collect breast milk from mice, a new approach from previous data collection techniques. Moreover, another one of her research projects led to the addition of 2’-fucosyllactose (Figure 1), a sugar present in breast milk, to formula for full-term babies because of its ability to protect infant intestines.3
Dr. Good continues to pursue
her interest in combating intestinal disease through leading her lab in the exploration of NEC from every direction as they aim to work towards finding a cure. The lab has determined potential preventative strategies, such as compounds found in green leafy vegetables that improve the intestinal environment, specifically the epithelial barrier, and lower inflammation in mouse models of NEC.2 In addition to studying the prevention of NEC, the Good Lab aims to fight back against the disease. Previous research in adult disease models revealed that interleukin 22 (IL-22), a molecule involved in the immune system, protected against injury and inflammation of the intestine.4 Inspired by these findings, Dr. Good investigated IL-22 in relation to neonatal intestinal health and demonstrated its promise as an immunotherapy for NEC. Treating neonatal mice, which serve as an experimental model of NEC, with IL-22 showed an increase in intestinal epithelial cell regeneration and less intestinal injury.4 On top of having a patent for the treatment of NEC with IL22, the Good Lab is working to advance the research to clinical trials in the future and continue studying what makes IL-22 protective. Dr. Good explains, “Figuring out how to harness the good part of the inflammatory response versus the bad is a really critical factor of our
lab.”2 Understanding the full protective abilities and downstream effects of IL-22 during NEC will be a huge step toward improving the treatment of infants suffering from the disease.
Once she entered the fight against NEC, Dr. Good quickly understood the value of collaborating with other researchers to study such a complex disease that has a relatively low prevalence at each hospital. Therefore, she founded the NEC Biorepository connecting NEC researchers nationwide (Figure 2). Currently, eight hospitals are enrolling patients, and it is the largest biorepository for patients with NEC in the United States.5 Scientifically useful samples, such as blood, urine, stool, saliva, resected intestines, and breast milk, are collected from preterm infants. These samples are used by the Good Lab and many other research laboratories to search for potential biological compounds, such as proteins or genes, that may indicate the potential for disease development and its severity in individual infants. The sample collection performed by the Good Lab is typically non-invasive, since primarily samples being discarded, such as urine and stool, are used, thus limiting the stress induced on preterm babies. Early identification of NEC risk can prepare medical professionals to try to prevent irreversible damage and save patient lives.
Another avenue of exciting research tackled by the Good Lab is the development of treatments for NEC that can be used in patients. After a potential therapy is discovered in the lab, typically the next step is to test it in animal models and then progress to trials with


human subjects. The Good Lab has worked to support the transition from animal to patient studies by inventing a model using intestinal epithelial cells grown from the tissues of infants.2 These tissues are obtained from patients when a part of the intestine needs to be removed from an infant to benefit their health, like if they have NEC or an intestinal obstruction. A small piece of the intestine is grown into what is called a “mini-gut”. The cells can then be put on a device called a microfluidic chip to grow and become an “Intestine-on-aChip model” that mimics the structure of the intestine of infants (Figure 3). Subsequently, introducing intestinal bacteria from a NEC patient to the intestinal model creates the “NEC-on-aChip” model, which allows the disease to be studied and potential therapies to be

evaluated prior to human trials. The progress of Dr. Good’s lab in analyzing NEC prevention and treatment has been extensive and inspires future work to fight the disease. Whether it be through methods for early diagnosis or immunotherapeutic compounds, highly innovative thinking will be needed to protect infants against this fast-acting disease. As long as those involved in the fight against NEC embody the mentality of the Good Lab through remaining inquisitive, embracing the unexpected, and approaching problems from a multitude of angles, there is hope for the eradication of NEC.
1. Lanik, W.E., Luke, C.J., Nolan, L.S., Gong, Q., Frazer, L.C., Rimer, J.M., Gale, S.E., Luc, R., Bidani, S.S., Sibbald, C.A., et al. JCI Ins. 2023, 8.
2. Interview with Misty Good, M.D., M.S. 2/12/24.
3. Good, M., Sodhi, C.P., Yamaguchi, Y., Jia, H., Lu, P., Fulton, W.B., Martin, L.Y., Prindle Jr., T., Nino, D.F., Zhou, Q., et al. Brit. Journ. Of Nutri. 2016, 116, 1175-1187.
4. Mihi, B., Gong, Q., Nolan, L.S., Gale, S.E., Goree, M., Hu, E., Lanik, W.E., Rimer, J.M., Liu, V., Parks, O.B., et al. Cell Rep. Med. 2021, 2.
5. Ralls, M.W.; Gadepalli, S.K.; Sylvester, K.G., Good, M. Semin. Pediatr. Surg 2018, 27, 25-28.

Diagnosed in 1 in 59 children in the United States alone, Autism Spectrum Disorder (ASD) is a neurological disorder associated with various levels of social communication deficits and restricted or repetitive behaviors and movements. While behavioral markers and profiles of motor abilities are typically assessed by individuals such as teachers, pediatricians, and parents, these markers are less noticeable in infants, and the average ASD diagnosis by a pediatrician or clinical child psychologist is made around the age of four. However, extensive behavioral and cognitive research has associated early intervention with better outcomes for children with ASD. This is where Dr. Jessica Girault’s research comes in.

With a Ph.D. in Neuroscience from the University of North Carolina at Chapel Hill, Dr. Girault was interested in studying the cognitive development of children. Specifically, she was curious about the progression and maturation of infantile cognitive abilities from complete reliance to speaking hundreds of words per minute. During her postdoctoral years, she began to specialize in the processes of the brain that went awry for infants diagnosed with ASD. Rather than analyzing standard behavioral and motor coordination markers, Dr. Girault’s lab explores ASD via
the emerging process of neuroimaging and machine learning. Neuroimaging is currently the most powerful, non-invasive imaging tool for individuals of all ages that measure the structure and function of one’s brain through white matter pathways. The primary goal of the Girault Lab is “to lower the age of Autism detection and develop interventions that are useful during an infant’s early window of cognitive development.”1
“Dr. Girault’s lab will revolutionize understanding, lower the age of diagnosis, and ultimately apply the proper intervention accurately and early.”
Because certain brain features are not detectable to the naked eye, neuroimaging captures images between different regions of neurons and anatomical structures to fully assess the brain’s pathology and structural components. These functional magnetic resonance imaging (fMRI) images have demonstrated great potential for information analysis and predictive abilities. Coupled with these fMRI images, the Girault Lab trains machine learning systems to improve their accuracy in assessing diagnoses for ASD. The machine learning systems identify images and output diagnosis results based on data sets; consequently, the Girault Lab has effectively trained computers to have an 80 % positive predictive value for unseen data.¹ According to Dr. Girault, this revolutionary method for diagnosis well before behavioral symptoms are present will “allow families to be
proactive and assign early intervention.”¹
UNC-Chapel Hill is one of the first universities in the country to focus on fMRI scanning for infants and numerous longitudinal studies have been conducted here since the early 2000s. Dr. Girault is involved in projects such as the Baby Connectome Project and Infant Brain Imaging Study (IBIS). Due to the genetic nature of ASD, her work at IBIS focuses on how infant siblings with autism put siblings at an exceptionally high 1 in 5 risk for diagnosis; her research follows infants from 2-6 months of age and measures fMRI differences before symptoms are detectable at around 18 months. Dr. Girault also works with Dr. John Gilmore from the UNC Department of Psychiatry in longitudinal studies of early brain development, as UNC-Chapel Hill is a central hub for infant brain development research.
As one might imagine, these research processes cannot be completed by neuroscientists alone — an interdisciplinary team of radiologists, engineers, and computer scientists is required for a variety of experimental procedures. Lab processes involve expertise from a variety of disciplines, and Dr. Girault emphasizes the crucial value that her interdisciplinary team provides in their respective roles. For instance, radiologists use medical image analysis to verify that the correct image is being analyzed; engineers and computer scientists help translate the code of the machine learning operating systems; behavioral psychologists assist in measuring overall infant cognition; graduate students and statisticians analyze and interpret data collection. Girault appreciates the crucial value that her interdisciplinary team provides in their respective roles.
As expected, a major research endeavor such as this often crosses numerous obstacles. Specifically, Dr. Girault highlighted the role of “Baby Whisperers” who are responsible for creating a comfortable, and relaxing environment for infant
study participants, some of whom are as young as six weeks. A major challenge, she noted, was navigating creative ways for families and infants to feel a sense of comfort during the scanning procedures. Additionally, as the lab rescanned the children at adolescence, the findings from these longitudinal studies challenged the accepted definitions of what constitutes autism and if this knowledge of brain development can be used to reevaluate autism.
Looking towards the future, Dr. Girault aspires to replicate machine learning papers and ensure robust data collection to justify the use of neuroimaging and machine learning. Presently, she is collecting a new cohort of about 180 infants to validate past results with a larger, scalable sample size with the ultimate goal to “solidify machine learning algorithms and predict whether they will develop ASD based on MRI and the use of randomized control trials.”2 This new cohort is part of emerging work analyzing sensory input even before birth; since brain circuits are primed for sensory information, Dr. Girault is interested in analyzing the integration of sensory and visual processing of patients with ASD. She defines this developmental cascade of her future work as the “difference in visual system attention to downstream social behavior and social learning and processing during the first year of life.”² With future understanding of the neurobiology of ASD even before birth, one can only imagine how Dr. Girault’s lab will revolutionize the scientific community’s understanding of autism in infants to ultimately lower the age of diagnosis and apply proper intervention accurately and earlier on.

Technology Networks.
References
1. Interview with Dr. Jessica Girault 2/21/24.
Imagine this scenario: a hot summer day, a lemonade stand, sticky dollar bills. Sounds like a custom-made invitation for fruit flies, especially if the lemonade stand is set up every day to make enough cash for that new toy advertised on TV. Like many other organisms, fruit flies use a process called associative learning to connect unrelated stimuli in the brain, which is why your citrus beverage startup is garnering publicity from the wrong species.
Learning and memory cause intricate changes in synapses, tiny gaps between neurons that allow for communication, and adjust their connectedness based on levels of neural activity. The highly adaptable nature of neurons is possible due to synaptic plasticity, a process critical to neural development and maintenance. Dr.
Toshihide Hige, assistant professor of Cell Biology and Physiology at the University of North Carolina at Chapel Hill, studies the mechanisms of synaptic plasticity in the fruit fly, Drosophila melanogaster. Dr. Hige’s research focuses on identifying neural circuits— connections between neurons responsible for a specific function. To examine how the activity of specific circuits corresponds with Drosophila behavior, the Hige Lab uses experimental techniques such as electrophysiology, or the study of a cell’s electrical properties, and behavioral assays.1 In a recent collaboration with researchers from the Howard Hughes Medical Institute and

Columbia University, Dr. Hige and his team identified several neural circuits that allow for higher-order associative learning in Drosophila 2

Figure 1. Illustration of Pavlov’s first-order conditioning experiment. (1) Unconditioned stimulus, food. Unconditioned response, salivation. (2) Neutral stimulus, bell. (3) Conditioned stimulus, bell. Unconditioned stimulus, food. Unconditioned response, salivation. (4) Conditioned stimulus, bell. Conditioned response, salivation. Image courtesy of Maxxl via Wikimedia Commons.
To study associative learning, scientists employ classical conditioning, a behavioral protocol that examines how specific stimuli are associated with punishment or reward. In first-order conditioning, the most basic form of classical conditioning, a neutral stimulus is paired with an unconditioned stimulus that produces an innate response; over multiple trials, the neutral stimulus is associated with the innate response. A common example is the Pavlovian dog experiment, in which the sound of a bell triggers salivation after being repeatedly presented with food (Figure 1).
Second-order conditioning is a higher-order form of associative learning that involves an additional step: A new stimulus is paired with a previously conditioned stimulus to evoke the same behavioral response, without direct exposure to the unconditioned stimulus. For instance, a cat previously conditioned to associate the sound of a can opener with food undergoes second-order conditioning to associate a squeaky cabinet door with food (Figure 2).
In the Drosophila brain, the mushroom body is a small structure important for olfactory learning and memory. A high concentration of neurons
that release dopamine, a neurotransmitter associated with the reward system and known to facilitate learning and memory, is present in the structure. According to Dr. Hige, the mushroom body is where the conditioned stimulus and unconditioned stimulus “meet” in the brain and is essential for classical conditioning to occur.3 Conveniently, most of Drosophila’s olfactory neural circuits are located in the mushroom body, where there are 15 segregated compartments. Each compartment contains neurons that release different neurotransmitters and is responsible for storage of different types of memory, which include short, long, aversive, or appetitive memory. Furthermore, the collective effort of the scientific community to study fruit flies offers a valuable well of information and resources, especially the extensive tools available for genetic manipulation; also, a “wiring diagram” for the entire Drosophila nervous system known as connectome has been mapped out, allowing scientists to target neural circuits more readily.3 These aforementioned characteristics make the mushroom body an exceptional model for studying the mechanisms of synaptic plasticity.

While the olfactory neural circuitry for first-order memory was well-studied in Drosophila, no research had explicitly identified second-order memory circuits before Dr. Hige’s team published their study. “In any animal model, the secondorder memory is always short-lasting compared to the first-order memory, no matter how many times we repeat [the protocols],” Dr. Hige said. “This makes it really difficult to dig into the circuit mechanisms.”3
The standard protocol for firstorder conditioning involves presenting a neutral stimulus with a sugar reward for an extended amount of time to invoke a desired response. However, secondorder conditioning forms an indirect association between a new stimulus and a desired response, linked only by an initially conditioned stimulus. Dr. Hige and his team found that second-order memory decayed within a day and was highly prone to extinction.2 The key to retaining the second-order memory was to establish a strong association between the previously conditioned stimulus and the reward before administering the second neutral stimulus.3 The Hige Lab established a robust protocol for secondorder olfactory conditioning, a significant achievement that lays the groundwork for future research in higher-order associative learning in Drosophila Using connectome data, Dr. Hige and his team identified and stimulated dopamine neurons of interest by employing a technique called optogenetics, which uses genetic drivers to activate neurons with light. They

also performed electrophysiological experiments using in vivo whole-cell patch-clamp recordings, which measures the electrical changes of neurons, in response to odors and light stimulation. To stimulate the neurons, the Hige Lab administered sugar rewards and used a modified version of the protein channel rhodopsin to excite specific neurons with red light. They targeted distinct sections of the mushroom body and found that the α1 compartment, known for longlasting appetitive memory, was most likely responsible for storing the firstorder memory that “instructs” secondorder conditioning.2
However, the α1 compartment, like all other structures of the brain, cannot do much without help from interneurons, which connect neurons with different functions and allow them to communicate. The Hige Lab identified a single key interneuron called SMP108 that connects the α1 compartment to dopamine neurons of other compartments (Figure 3). They blocked SMP108 with a neurotoxin and genetic drivers, which significantly decreased second-order memory scores, and concluded that it is required for secondorder conditioning. They also found that SMP108 allows for the formation of second-order memory by promoting dopamine release in multiple mushroom body compartments.2
As Dr. Hige and his team continue to probe the mysteries of the Drosophila brain, their discoveries will not only advance our understanding of neuroscience, but also inspire future research examining mechanisms for memory and learning in higher organisms, including humans. Uncovering new neural circuits might not prevent fly-infested lemonade stands or cure Alzheimer’s for now; however, these findings contribute to a deeper understanding of the fundamental principles of brain function and highlight the importance of studying simpler model organisms like fruit flies.
1. The Hige Lab. https://www.higelab. org/ (accessed February 20th, 2024).
2. Yamada, D.; Bushey, D.; Li, F.; Hibbard, K.L.; Sammons, M.; Funke, J.; Litwin-Kumar, D.; Hige, T.; Aso, Y. eLife 2023. 12:e79042.
3. Interview with Toshihide Hige, Ph.D. 02/05/24.

At an age often fraught with immense self-doubt and stress, laughter, as the old adage suggests, is often a welcome treatment for emotional ailments; however, it is not the best option out there. Through new research, mindfulness and selfcompassion have emerged as new frameworks with which young adults can orient their expectations of both the world around them, and most importantly, themselves. In one of her most recent publications, Dr. Karen Bluth explores this idea through a novel 6-week program known as Embracing Your Life: An Online Mindful Self-Compassion Program for Emerging Adults. Through studying the initial implications of the program, Dr. Bluth, along with her colleagues, were able to successfully demonstrate the program’s ability to significantly decrease depression, loneliness, and stress in young adults. With such encouraging results, Dr. Bluth and her colleagues are working diligently to expand the program both domestically and internationally to use the ideas of mindfulness and selfcompassion to help young adults around the globe in their pursuit of happiness.
Dr. Bluth began her journey of helping teens and young adults early,
spending time as a middle school educator before returning to school for her PhD. It was during her time as an educator that she noticed how deeply teens and young adults were struggling, especially with novel sources of adversity such as the rise of social media. She notes how she loves working with that age group as “they have so much to offer” and are really “open, interested, and articulate” in discussing their struggles in the right setting.1 Throughout the course of her time at the University of North Carolina at Chapel Hill, Dr. Bluth has spent more than a decade researching mindfulness and self-compassion in adolescents with the goal of helping to reduce the prevalence and severity of ailments such as depression and anxiety in teens and young adults alike. Her most recent endeavor in pursuit of this goal, the Embracing Your Life program, has offered a route to do that.
Core to the Embracing Your Life program are the ideas of mindfulness and self-compassion. The former, defined simply as an understanding and awareness of what one is feeling as they feel it, is an integral component of the latter, and something Dr. Bluth believes is essential in promoting a healthy mental
state. As defined in the foundational research by Dr. Kristin Neff that Dr. Bluth cites, self-compassion, and her program by extension, has three essential components: mindfulness, common humanity, and self-kindness.2 The first component, mindfulness, centers around not catastrophizing distressing events and acknowledging the associated negative emotions, instead of “shoving them under the rug”.1 The second component, common humanity, is rooted in the understanding that difficult
 Karen Bluth, PhD
Karen Bluth, PhD

1. The Embracing Your Life program teaches participants how to be their own best resource when they are struggling. Image courtesy of Wikimedia Commons.
feelings are part of what we experience as humans, and that disruptive negative emotions do not imply an inherent fault, but rather, indicate a common human nature. Dr. Bluth emphasizes how this component has been especially difficult in recent years given the prevalence of social media and the associated popular false narratives. Correspondingly, a section on social media was specifically added to the Embracing Your Life program due to its well-known role as a metric for negative comparison for many young adults. The common humanity piece is also a large reason Dr. Bluth conducts the Embracing Your Life program in groups. Participants cited that seeing that they are not alone in their struggles, that they are not an aberration, was integral in adopting a more self-compassionate mindset. The final component, selfkindness, is the physical manifestation of the first two and involves taking an “active step to support yourself” by recognizing what one needs at the moment and doing it.1 This component can take many forms, from as simple as saying kind words to oneself or taking a walk to more involved things such as talking to a friend or finding a new hobby. Applying these three components, Dr. Bluth and her colleagues built the Embracing Your Life program to enable participants to utilize guided practices and supportive gestures stemming from the core components of self-compassion in order to support themselves when they are struggling.
The program was first applied in 2020 by Dr. Bluth and her colleagues, then later analyzed in Feasibility, Acceptability,
and Preliminary Outcomes of Embracing Your Life: An Online Self-Compassion Program for Emerging Adults, published in 2023. The publication describes the initial implication of the study in 2020 during the COVID-19 pandemic.3 Dr. Bluth and her colleagues knew the program could help many people and wanted to support its effectiveness with research. However, research funding often takes a very long time, so they worked without funding for the duration of the study, essentially volunteering their time during the pandemic to institute and improve the program to help those they could. Although the additional isolation significantly increased feelings of loneliness and depression universally, Dr. Bluth and her colleagues have done studies, even encompassing a wider age range, both before and since the pandemic that yielded similarly positive results. She points out how, in our society, people tend to look to others for support. While socialization is a great tool to improve mental health outcomes, the ability to be “your own best friend” is indispensable, especially during isolating events such as COVID-19.1
From Europe to South America to Asia, through the Embracing Your Life program, Dr. Bluth has been able to reach young adults around the world. In the effort to expand the program, beginning

this Spring, Dr. Bluth and her colleagues will begin training educators to utilize the program to foster self-compassion to a similarly international extent. Despite its daunting scale, the process is not a novel one for Dr. Bluth as, in the past, she has taught international educators a similar program built for teenagers with great success in different countries. She describes how, although they adapt the curriculum for different cultures, they do not see significant differences in outcomes regardless of where it is taught. In addition to training, Dr. Bluth and her colleagues are presently working on publishing her methods in a new book to further expand their reach. Regardless of the medium, the results are clear, selfcompassion is something the world needs more of, a sentiment increasingly true for young adults. To that end, Dr. Bluth and her colleagues work every day to get the program to as many young adults as it can help, because, as she describes it “the more, the better”.1
1. Interview with Karen Bluth, Ph.D. 2/9/2024
2. Neff, K.; Self-Compassion: Theory, Method, Research, and Intervention; Annual Review of Psychology 2022, https://doi.org/10.1146/annurev-psych-032420- 031047.
3. Bluth, K.; Knox, M.; Press, A.; Lathren, C.; Feasibility, Acceptability and Preliminary Outcomes of Embracing Your Life: An Online Self-Compassion Program for Emerging Adults; Sage Journals 2023, https://doi. org/10.1177/216769683231189902.

 Dr. Tom Rodebaugh
Dr. Tom Rodebaugh
differs from simply being afraid of talking to people and goes into a further fear and sometimes even paralyzing debilitation in social situations. Many studies have investigated how best to treat SAD with the most common conclusion being to face your fears. This means that instead of trying to avoid a situation that would cause anxiety, people with SAD are encouraged to engage in those situations. Facing your fears has been the most important part of Cognitive Behavioral Therapy which is a treatment that helps to break down overwhelming situations into parts or smaller steps to help them be more manageable, it is the most widely used treatment for SAD. Previous studies, specifically randomized controlled trials, have promoted the theory that
However, some professors have found that treatment may be more complicated than that, Dr. Tom Rodebaugh being one of them.
Dr. Rodebaugh is a new psychology professor at the University of North Carolina at Chapel Hill this year from Washington University.3 He graduated with a masters and PhD in Psychology from UNC after completing his bachelors at Pennsylvania State University. Dr. Rodebaugh started his work with anxiety research while at UNC with Dr. Dianne Chambless who introduced him specifically to the topic of social anxiety and he found that it fit his interests and his professional capabilities very well. In the past few years of his work, he has been excited to be able to “use a lot of tools that just weren’t available 10 to 20 years ago to answer questions that we couldn’t even figure out to ask in the past.” Dr. Rodebaugh’s transition to the University of North Carolina has also given him the chance to work with other psychologists in the field who have similar interests to him.
day to day basis had any effect on social anxiety disorder the next day. Avoidance is defined, for this intent, as attempting to stay away from situations that would cause anxiety. The hypothesis, as established by previous studies, was that avoidance would increase the next day levels of anxiety felt by participants. The team completed a study on 32 participants with SAD over a period of 120 days in which each participant was provided with a survey throughout the day and asked to answer prompts about their anxiety levels and their levels of avoidance. These prompts included things like “Today I felt __ about social situations”2 and avoidance levels were self-reported by individuals on a sliding scale of what specifically they avoided and to what extent, whether that be social situations as a whole or even simply thinking about them. These surveys were considered the first part of the study, after 120 days if participants had completed 100 of the 120 studies, they were reassessed for SAD, given a speech to perform, and were asked to self-report their anxiety levels. After this,
participants had a brief period of time, typically five days, in which they would continue completing surveys before meeting and performing these same tasks a second time.2
After the study was completed, Dr. Rodebaugh and the team of researchers discovered that social anxiety did have a correlation with avoidance. That correlation was that “avoidance on one day did predict anxiety the next”.3 It was clear this evidence showed that avoidance maintained and increased fear for a small number of individuals as is consistent with previous research, however, there was not significant evidence that avoidance increases fear in all instances of SAD. The conclusion the team reached was that their “hypothesis that the strength of prediction between fear and avoidance would predict exposure response was
not convincingly supported. However, [they] saw preliminary evidence that the autoregression of social fear predicted exposure response.”2 Autoregression is the concept that past reactions will dictate and predict future reactions. This means that the team was able to show how people get stuck in loops of avoidance and anxiety and its connection to increased or stagnant anxiety the next day.
anxiety as it presents, there would be a clearer understanding of what therapies specifically help and what the connection of avoidance as a factor in increasing anxiety actually is. This study would be done specifically on speech anxiety as it is one of the subsets of social anxiety that was studied originally.
This research and likely more of Dr. Rodebaugh’s studies in the future could change how SAD is treated, hopefully for the better of many adults and teens who have this disorder. The new evidence suggesting that avoidance does not worsen anxiety on a day to day basis for everyone goes against most of what therapists are often taught to teach their clients. It is unclear just how much this will affect treatment but Dr. Rodebaugh’s future research into the aspects of cognitive behavioral therapy that truly help social anxiety disorder might provide a more direct approach to treatment that hasn’t been used much until now. If research continues to trend towards avoidance maintaining levels of anxiety in most individuals, it could incite a change in the direction cognitive behavioral therapy takes, possibly causing more psychologists and therapists to help patients find a better way to face their fear.
This study didn’t dispute that avoidance is increasing some people’s fears but it did show that some people have gotten to a point where “they avoid so much already that knowing about their day to day avoidance will not help predict”3 their anxiety in the coming days. To continue this line of study, Dr. Rodebaugh is involved in developing a similar study with the added condition of attempting to help people with their anxiety while the study is going on. The goal would be to observe what causes symptoms to get better during treatment, specifically different aspects of cognitive behavioral therapy. By treating the
1. Social anxiety disorder. Anxiety and Depression Association of America, ADAA. (n.d.). https://adaa.org/ understanding-anxiety/social-anxietydisorder
2. Rodebaugh, T. L., Grossman, J. T., Tonge, N. A., Shin, J., Frumkin, M. R., Rodriguez, C. R., Ortiz, E. G., & Piccirillo, M. L. (2024). Avoidance and fear day by day in Social Anxiety Disorder. Psychotherapy Research, 1–14. https://doi.org/10.1080/10503307.2023. 2297994
3. Interview with Tom Rodebaugh
4. 13 reasons why gen Z stress about Social Anxiety. YouthSense. (2019, September 4). https://www.youthsense. com.au/parents/why-gen-z-stress-oversocial-anxiety
5. National Collaborating Centre for Mental Health (UK). (1970, January 1). Social anxiety disorder. Social Anxiety Disorder: Recognition, Assessment and Treatment. https://www.ncbi.nlm.nih. gov/books/NBK327674/

In the realm of cutting-edge neuroscience, Dr. Dea Garic at the University of North Carolina at Chapel Hill has embarked on a journey to unravel the mysteries surrounding the early markers of autism spectrum disorder (ASD). Through a longitudinal study examining infants from 6 to 24 months of age, her work has focused on the intriguing connection between enlarged perivascular spaces (PVS) in the brain and their link to an early autism diagnosis, cerebral spinal fluid (CSF) volume, and later sleep problems.

Notably, Dr. Garic's research has uncovered a correlation between enlarged PVS, increased CSF volume, and the emergence of sleep issues, marking these spaces as potential early biomarkers for autism. These fluid filled spaces play a crucial role in cleaning the brain during sleep. Strong waves of CSF wash through our brains as we slumber, flushing out waste products and toxins, a process vital for proper development
and cognitive function.1 Ehnlarged PVS are thought to indicate that this flow of fluid is stagnant, and the absence of this cleaning process can lead to developmental delays and cognitive deficits, emphasizing the importance of understanding the link between an enlarged PVS and neurodevelopmental disorders such as autism.1, 2, 3, 4

1. Extra-axial cerebrospinal fluid (EA-CSF) and perivascular space (PVS) segmentation on 24-month scan of a child who was diagnosed with autism. Image courtesy of Dr. Dea Garic.
Expanding on her background in white matter tractography, a technique used in neuroscience to map the pathways of nerve fibers in the brain, Dr. Garic and her team have examined infant MRIs at various stages, taking snapshots of brain development from 6 to 24 months. The strength of their approach lies in non-invasive imaging techniques. “Some individuals with autism don’t even get diagnosed until adolescence because they have a completely separate phenotype,” she notes.5 Imaging techniques, however, allow for a detailed exploration of the neural landscape
without posing any harm to the subjects, allowing for a more comprehensive examination than traditional diagnostic methods. Her findings of enlarged PVS in infants challenge previous assumptions about these spaces and their role in brain health. As individuals age, perivascular spaces naturally enlarge. Neuroradiologists are familiar with this phenomenon but were surprised and intrigued when this enlargement was visible in infants and toddlers.
This early identification of enlarged PVS could have significant implications for early intervention and treatment of neurodevelopmental disorders, marking a significant advancement in radiology.
“My original interest was to find biomarkers in the brain…I became fascinated with the idea of finding biomarkers and biological mechanisms that might precede clinical outcomes,” she explains. “Our overall goal is to be able to use neuroimaging findings to

Figure 2. CSF normally flows continuously throughout the perivascular spaces, where it filters out inflammatory proteins. New findings now suggest that nearly half of infants who go on to develop autism have enlarged PVS by two years of age. Image courtesy of Dr. Dea Garic.
predict clinical outcomes.” 5
Considering the diverse subtypes of autism, the goal is to use these findings to predict disorder severity and provide targeted interventions. By understanding the association between enlarged PVS and clinical outcomes, the research aims to provide insights into clinical issues that parents and children often face. For example, sleep disturbances are prevalent in children with autism, and early detection could lead to interventions that improve sleep quality and overall development.
“Early brain markers when the mom is pregnant…that is the natural next step of the future,” she reflects.5 However, she raises important bioethical questions about whether parents truly want to know about potential developmental concerns so early in infancy and if there are genuine advantages to such early revelations. Bioethics is an important consideration in the field of clinical neuroimaging and one that the Infant Brain Imaging Study (IBIS) Network is continuing to examine, raising important questions about the implications of early diagnosis for parents and infants. While early detection of potential developmental concerns can lead to timely interventions and support, it also presents challenges. Parents may be faced with difficult decisions and emotional distress upon receiving such information about their child's future, posing a complex ethical dilemma. Additionally, while early diagnosis may offer advantages in terms of early intervention and improved outcomes, it also raises concerns about labeling and
stigmatization. As Dr. Garic's research continues to reveal new insights about early biomarkers for autism; bioethics will play a crucial role in guiding the ethical implementation of procedures resulting from these discoveries.
The journey into the neural landscape has significant clinical implications for formerly established practices. The surprising discovery of these enlargements at such an early stage challenges traditional assumptions about brain development, which could impact how routine checks are conducted. In conjunction with IBIS, sprawled across five sites in the United States, Dr. Garic acknowledges that data acquisition and organization has not been a major issue. She plans to continue collaborating with IBIS and neuroradiologists, potentially scanning the same cohort of infants in adolescence to explore the long-term implications of enlarged PVS and assess symptom severity. Additionally, she is interested in collaborating with other large networks, such as the Human Connectome Project (HCP), to delve deeper into the biological mechanisms underlying these intriguing phenomena. The HCP is a large-scale research project that has tackled one of the greatest challenges in recent scientific exploration: mapping the human brain; aiming to connect its structure to function and behavior. This project has generated a wealth of data that is available to the scientific community, enabling researchers to study the brain’s neural pathways and structure in unprecedented detail.6 Dr. Garic's interest
extends beyond autism, as she is also “interested in examining whether these signs of glymphatic dysfunction also exist in other genetic syndromes,”5 hoping to uncover shared physiology between autism, and other neurodevelopmental disorders such as Down Syndrome. Dr. Dea Garic's research on enlarged perivascular spaces in infancy and their link to autism represents a significant advancement in our understanding of early biomarkers for autism spectrum disorder. By unraveling these early signs, Dr. Garic's work offers hope for improved early detection and targeted interventions, ultimately aiming to enhance the lives of individuals affected by autism and their families.
1. Garic, D; McKinstry, R. C.; Rutsohn, J. Enlarged Perivascular Spaces in Infancy and Autism Diagnosis. JAMA Network Open. 2023, 6(12). DOI: 10.1001/ jamanetworkopen.2023.48341
2. Kern, K. C.; Nasrallah, I. M.; Bryan, R. N; Reboussin, D. M; Wright, C. B. Intensive Systolic Blood Pressure Treatment Remodels Brain Perivascular Spaces: A Secondary Analysis of the Systolic Pressure Intervention Trial (SPRINT). Neuroimage Clin. 2023, 40. DOI: 10.1016/j.nicl.2023.103513
3. Xie L, Kang H, Xu Q, etal. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156): 373-377. doi:10.1126/science.1241224
4. Wardlaw J.M., Benveniste H, Nedergaard M, et al. Perivascular spaces in the brain: anatomy, physiology and pathology. Nat Rev Neurol 2020;16(3):137-153. doi:10.1038/s41582020-0312-z
5. Interview with Dea Garic, Ph.D. 01/23/24.
6. Van Essen, D. C.; Smith, S. M.; Barch, D. M.; Behrens, T. E. J; Yacoub, E; Ugurbil, K. The WU-Minn Human Connectome Project: An Overview. Neuroimage 2013, 80, 62-79. DOI: 10.1016/j.neuroimage.2013.05.041
7. Enlarged Spaces in Infant Brains Linked to Higher Risk of Autism, Sleep Problems. UNC Health Newsroom, 2023. https://news.unchealthcare. org/2023/12/enlarged-spaces-in-infantbrains-linked-to-higher-risk-of-autismsleep-problems/ (accessed 2024-03-19).

It is commonly thought that memories freeze in time, captured forever like a picture. However, this is usually not the case; memories are often adapting as we continue to change minor details that occurred in the memory. Not only does the content of our memories change but our emotions attached to that memory can change over time as well. The phenomenon, known as memory modification, is the potential to change events that occur in a memory as well as feelings attached to that memory before reactivation.1 For example, one may have a memory of a bad experience of seeing a snake, over time their negative emotions may decrease towards the snake because they cannot fully recall their feelings during the experience.3 Memory modification is used unintentionally in daily life, but what if there was a way to use memory modification to change negative feelings towards unpleasant memories? Dr. Natasha Parikh, a professor at the University of North Carolina at Chapel Hill’s Department of Psychology and Neuroscience, researches how spatial distancing—spaced out intervals—is
The phenomenon, known as memory modification, is the potential to change events that occur in a memory as well as feelings attached to that memory before reactivation.
used to reduce negative emotions that may arise when reactivating certain memories. Her research includes testing emotional arousal towards negative memories over a period of time to determine whether reactivation across periods of time reduces emotional arousal by the modification of the memory.3
Dr. Parikh’s study consisted of 128 participants in North Carolina and participants were asked to take part in three different sessions throughout the study.2 There are two different components of spatial distancing: reactivation and regulation. Reactivation refers to when memories resurface and regulation refers to the control of emotions. Participants were split into four different groups: reactivation+regulation (experimental group), no reactivation (control for the regulation group), no regulation (control for just reactivation) and neither (overall control).2 Using these four different groups allowed for the researchers to determine if there was a particular combination
of the methods that produced the least emotional arousal to the images. During the first session, participants were given 25 different images and were asked to rate the images based on their arousal (1=calm to 9=excited). The images were chosen by using the International Affective Picture System (IAPS) which allowed the researchers to choose images that had similar ratings for arousal.2 In addition, using this system allowed for the researchers to ease participants into looking at negative images. For session two, the groups took part in different activities in order to measure how important each individual aspect of spatial distancing was to the

reduction of emotional arousal. The reactivation+regulation group was asked to view emotional objects cut out from the test pictures and then asked to regulate them and rate the success of their implementation from a scale of one to four. The no-reactivation group followed the same emotional regulation procedures as the reactivation+regulation group but instead of the test images which included the emotional objects/ aspects, participants saw control images in which the emotional aspects were cut out. The no-regulation group was presented the same test images as the reactivation+regulation group, but instead of rating their successes at regulation, they were asked if the subject portrayed a human or not, which was used as a judgement control. After the first two sessions, participants were asked to wait 96 hours until they came in for the third session in order to allow them to fully process the images.2 During the third session, participants were given the original test images as well as a mix of new images and were asked to view the image for three seconds, then rating the images based on valence and arousal they caused. They were then asked if the image they had seen was an old image or a new image and rated how certain they were on a scale from one to four.
After each session, participants were required to fill out the “Subjective Units of Distress Scale” to assess distress levels that may have risen after seeing the images. In addition, emotional arousal was measured through looking at the participants sweat levels using electrodermal activity measurement to see how participants responded to seeing the images throughout the three sessions. To account for any confounding factors that may have contributed to the response of participants, they were given surveys concerning their susceptibility to response bias before the study. The Social Desirability Scale questionnaire was given to participants beforehand to test their propensities to give desired responses which allowed the researchers to determine how much of their answers were due to social desirability.
Through the usage of ANOVA testing to test the significance of emotion regulation and memory reactivation in relation to spatial distancing, it was found out that the combination of reactivating and regulating the memory significantly

Figure 1. This figure depicts the experimental design of the study and depicts the four groups of participants used in the study. The images shown in the figure are examples of the types of images that could have been shown to participants. Image courtesy of Dr. Parikh.
reduced emotional arousal compared to just reactivation or just regulating. Participants who participated in both the reactivation and the regulation had the strongest reduction in arousal. This shows that distancing techniques are very impactful in the reduction of arousal after the reactivation of memories.
The usage of spatial distancing to reduce the negative effects that come with unpleasant memories can be used in many different clinical settings such as helping people through phobias and helping people who have PTSD.3 Applications of this research can provide people with healthier ways to process unpleasant memories and can help them to overcome certain fears that have been a result of negative memories.3 The use of intentional memory modification
through spatial distancing is an incredible phenomenon that can be used to help people process their experiences.
1. Kredlow, M. A., Eichenbaum, H., & Otto, M. W. (2018). Memory creation and modification: Enhancing the treatment of psychological disorders. The American psychologist, 73(3), 269–285. https://doi.org/10.1037/amp0000185
2. Parikh N, McGovern B, LaBar KS. Spatial distancing reduces emotional arousal to reactivated memories. Psychon Bull Rev. 2019 Dec;26(6):1967-1973. doi: 10.3758/s13423-019-01648-z. PMID: 31385205.
3. Interview with Natasha Parikh, Ph.D. 2/09/2024

Babies may be trying to tell us something. Luckily, researchers have been observing behavioral patterns in infants as predictors for possible conditions later in their lives. Attention deficit hyperactivity disorder (ADHD) does not fall short of being one of the many disorders that have been researched in children, adolescents, and adults. However, there seems to be a research gap between early symptoms in infants and the diagnosis of ADHD later in life. Dr. Rebecca Stephens tackles this gap by providing research that dives into the connection between infant attentional behaviors and early ADHD symptomatology. Dr. Stephens is a research assistant professor in the Department of Psychiatry at the University of North Carolina at Chapel Hill with a research focus on developmental psychology, mainly focusing on cognitive development in early childhood.

Researchers have been able to pinpoint predictors for certain behaviors or cognitive issues early in age. ADHD is one of many neurodevelopmental disorders in children that has been researched extensively to discover predictors and symptomatology. Neurodevelopmental disorders, such as ADHD and autism, are categorized as conditions that are caused by impairment in areas such as cognition or behavior.1 “Theoretically, early attention-based behaviors are related to a number of behaviors later on.” Dr. Stephens points out.2 One common symptom often observed in children is being characterized as being highly distracted or unable to
focus attention for extended periods of time. However, this symptom or behavior is often recognized in middle childhood (between 6-12 years old) rather than in early childhood (2-8 years old) or infancy.
With limited research in this area, it is often difficult to pinpoint whether indicators for limited attention in infancy can be linked to ADHD in middle to later childhood. One significant indicator of ADHD that is often researched is executive function (EF). Executive function can be defined as the mental processes required to concentrate and pay attention to mental goals and tasks.1 Executive function is a key aspect of ADHD in terms of allowing one to achieve goals and remain focused on specific tasks presented to them. Oftentimes when looking in at children and adults, EF deficits are a key aspect of symptomatology.
But how are we tackling the issue of looking at possible links between attentional behaviors, executive function, and ADHD symptoms in infancy? This is a question that Dr. Stephens sought to answer. “There is not a ton of research that looks at attention in infancy and how that relates to ADHD later on,” Dr. Stephens mentioned when discussing the goals of her research.2 The goal of this research was to provide a first look into the possibility of finding any early signs or symptomatology of ADHD in preschool children and infancy that may indicate the possible diagnosis of ADHD in the future.
FYIv2.0 is a measure that was designed by a group of UNC researchers over a decade ago, which focused on flagging infants for a diagnosis of autism. FYIv2.0 contains a database of infants who have been previously flagged by the measure and whether they were later diagnosed with autism or not diagnosed. Furthering the research of FYIv2.0, Dr. Stephens and her team were interested in exploring other possibilities of using this measure to flag for other neurodevelopmental disorders other than autism. There are overlapping symptoms between

1. Configuration of FYI constructs highlighting what is being observed in each attention-based construct NSA, RSA, and ISA. Figure courtesy of Stephens et al.
autism and ADHD, so the possibility of flagging ADHD using the FYIv2.0 construct originally used for autism could be possible.
Using the FYIv2.0 measure, Dr. Stephens and her team created three attention-based constructs that categorized both the social and nonsocial characteristics of infant attention.3 By utilizing autism metrics, it may be possible to determine if there is also a correlation with later ADHD diagnosis. These constructs allowed them to draw connections between the FYI2.0 measure that focused on flagging for autism and the possibility of using them for ADHD diagnosis. The Attention-based constructs consisted of Responding to Social Attention (RSA), Initiating Social Attention (ISA), and Nonsocial Sensory Attention (NSA).1
These constructs allow for the measurement of ADHD symptomatology in infants by observing the connection between executive function and attention. Figure 1 provides examples of behaviors or actions observed for each of the social constructs. Nonsocial Sensory Attention (NSA) refers to the ways a child acts in response to sensory features of objects or their own body. Nonsocial Sensory Attention can be observed as the child rocking their body back and forth or the baby repeating an activity over and over. Responding to Social Attention (RSA) refers to the child's response or delayed response to the adult's invitation of attention. This can include the child looking at the adult when their name is called or turning their head to look at an object that has been pointed out by the adult. Initiating Social Attention refers to a child actively seeking the attention of another individual in a social setting. Some examples of this can be drawing attention to themselves or drawing attention to an object of desire.
Dr. Stephens’s research found that there were significant associations with nonsocial sensory attention (NSA) in 12-monthold infants and 4.5-year-old children with executive function and ADHD symptomatology.1 Within the 3.5 age gap, there were significant findings that early infant attentional behaviors may be correlated to ADHD and executive function. Taking a deeper look at the FYIv2.0 measure, it can be deduced that sensory features within the NSA construct that were primarily
made to detect autism spectrum disorder (ASD) can be linked to similar behavioral sensory issues observed in ADHD. Results support that executive function deficiency may be related to early sensory behaviors as indicated in the NSA construct. Both executive function and NSA are related to working memory and inhibition.
With Dr. Stephens findings, early intervention serves as the next step in this research. With the pursuit of more research into infant attentional behaviors and ADHD, early intervention serves to use predictors earlier in development to encourage the introduction of resources to set children up for success. Although Dr. Stephens's research serves as a preliminary look into this area, with adequate studies it is possible to fill a gap within this area of cognitive developmental psychology. Babies may be trying to tell us something, and researchers are ready to pay attention!
1. Stephens, R. L.; Elsayed, H. E.; Reznick, J. S.; Crais, E. R.; Watson, L. R. Infant Attentional Behaviors Are Associated with ADHD Symptomatology and Executive Function in Early Childhood. Journal of Attention Disorders 2020, 108705472094501. https://doi.org/10.1177/1087054720945019.
2. Interview with Rebecca Stephens, Ph.D. 02/22/2024
3. Stephens, R. L.; Sabatos-DeVito, M.; J. Steven Reznick. The Development and Validation of Attention Constructs from the First Year Inventory. Psychological Assessment 2017, 29 (5), 568–581. https://doi.org/10.1037/ pas0000380.
Binge drinking is a common yet risky behavior among college students, often seen at parties, sporting events, or just for fun. However, many are unaware of the lasting effects that alcohol has on the brain. Dr. Fulton Crews, a pioneering neuroscientist and the founder of the University of North Carolina at Chapel Hill Bowles Center for Alcohol Studies (BCAS), has dedicated much of his career to exploring how binge drinking affects the adolescent brain.
Dr. Crews has always been fascinated by mental health and the potential to reverse brain diseases. After earning a bachelor’s degree in biology, his initial path led him toward a career in medicine, working at the Upstate University Hospital. However, this experience shifted his focus from practicing medicine to a more profound interest in the effects of drugs on the brain. Through this experience, Dr. Crews became fascinated by the impact alcohol had on the brain of intoxicated patients. Pursuing this passion, Dr. Crews obtained his Ph.D. in Pharmacology from the University of Michigan, which led him to the National Institutes of Health, where he worked under the guidance of Nobel Prize winner Julius Axelrod. During his career, Dr. Crews initially studied antidepressants, receptors, and neurodegeneration in Alzheimer’s disease and alcohol use disorder, among other neuroscience studies.1
Recognizing his contributions and expertise, the UNC School of Medicine asked Dr. Crews to focus on alcohol research and develop the BCAS. As its director for 28 years, Dr. Crews led groundbreaking research on alcohol’s impact on the brain until stepping down in 2022.3

Dr. Crews continues to lead the 10- 10-component consortium Neurobiology of Adolescent Drinking in Adulthood (NADIA). Through this research initiative, Dr. Crews collaborates with numerous colleagues across the country, working collectively to investigate the effects of adolescent alcohol exposure on adult brain functionality. The core aim of the NADIA is to test the hypothesis

Figure 1. Indomethacin restores the basal forebrain cholinergic neurons after Adolescent intermittent ethanol exposure (AIE) in both male and female rats. Image courtesy of Dr. Crews.
that underage drinking alters adolescent brain development causing long lasting changes in adulthood that likely persist for life. Within the NADIA consortium, researchers mimic binge drinking using the adolescent intermittent ethanol (AIE) rat model, involving alcohol administration in a pattern mimicking human weekend and holiday adolescent binge drinking. However, adolescence in humans lasts about ten years, with puberty in the middle, whereas in rats it lasts only about three weeks. Thus, in the AIE rat model, male and female rats are dosed with binge drinking doses body weight per day for two days on and two days off-cycle across puberty from postnatal day 25 to postnatal day 54. Rats are then allowed to mature to adulthood without further exposure to alcohol. Studies have found altered adult behavior, neurochemistry, and gene expression lasting until at least postnatal day 200. Using this model, AIE increases adult binge drinking (in humans, defined by five drinks in males and 4 in females who are smaller over the span of two hours) as well as perseveration-like cognitive deficits, increases in anxiety, and pain sensitivity. NADIA aims to find the mechanisms of binge drinking-induced persistent changes in adult behavior.3 Through the use of animal models, Dr. Crews and his colleagues have successfully replicated binge-like drinking

Figure 2. Indomethacin restores learning deficits in both male and female rats during the use of the Morris water maze task. Image courtesy of Dr. Crews.
behavior using the adolescent intermittent ethanol (AIE) model. This method allowed Dr. Crews to identify significant behavioral shifts in the rat model, including increased anxiety, elevated alcohol consumption, social interaction deficits, and cognitive impairments, leading to diminished behavioral flexibility as well as increased anxiety and pain sensitivity. Dr. Crews observed that the environment can affect gene expression, leading to increased immune signaling and a loss of cholinergic neurons following AIE exposure to ethanol. In the human brain, cholinergic cells are pivotal for cognitive functions, including learning, memory, and behavioral responses. These cells are integral to decision-making processes and arousal, underlining their importance in daily cognitive functions. A decline in cholinergic cell numbers can have detrimental effects on memory and learning capabilities. This reduction compromises the brain’s ability to process and regulate information. In cases of neurodegenerative diseases like Alzheimer’s and Parkinson’s, the significant loss of these neurons is associated with neuroinflammation, contributing to cognitive decline due to the diminished presence of cholinergic cells necessary for adequate brain function.
The harmful impacts of ethanol exposure on cholinergic cells parallel the deficits observed in Alzheimer’s and Parkinson’s diseases, indicating a concerning link between alcohol consumption and neurodegenerative conditions. Dr. Crews’ research discovered that alcohol can trigger changes in gene expression through epigenetic mechanisms due to neuroinflammation. Although cholinergic neuron loss was generally interpreted as nerve cell death, Dr. Crews wondered if reversing neuroinflammation medications could reverse these ethanol-induced changes.
Using the AIE model, Dr. Crews first found exercise could reverse the neuronal loss and later extended that to indomethacin, an anti-inflammatory drug. Dr. Crews wanted to test the hypothesis that using indomethacin could restore AIE-induced loss of adult forebrain cholinergic neurons. For this study, two cohorts of both male and female rats were dosed using the AIE model. AIE rats were alcohol-free for two weeks, during which one group received indomethacin or a placebo (vehicle). The first cohort of the indomethacin study was euthanized on the postnatal day, 70 days, and 26 days after the last ethanol exposure to determine cholinergic and proinflammatory immunohistochemical markers. Immunohistochemistry (IHC) enables researchers to identify specific proteins using antibodies,
allowing identification and quantification of cholinergic neurons and proinflammatory proteins. A second cohort was assessed for spatial learning and memory capabilities using the Morris water maze test. This assessment helped determine the rats’ ability to remember their environment, aided by various spatial cues. Over six days, the rats underwent the Morris water maze testing three times daily. They were placed in a water-filled pool with a hidden platform, which served as the metric for evaluating their spatial learning. Each trial started with the rats placed in different locations, compelling them to distinguish the platform’s location and recall its precise position. Rats learn with each trial, as indicated by more rapidly finding the platform. AIE did not impact the learning of the maze. The ability to learn new things is termed behavioral flexibility, and this is tested using a “reversal learning” test requiring rats, having already completed the spatial learning test, to adapt to the platform’s relocation, testing their ability to learn the new position of the platform.2
The findings from the study were remarkable. Upon examining the cohorts of female and male rats that were subject to immunohistochemistry (IHC) post-euthanasia, it was observed that those administered AIE experienced a significant decrease in choline acetyltransferase (ChAT), an enzyme used as a marker that is essential for producing neurotransmitters involved in learning and memory within the basal forebrain. ChAT serves as a marker for cholinergic cells and can impact learning, memory, and cognitive processes. As for the results, there was a 33% loss of ChAT in males and an 11% loss in females. Conversely, the cohort treated with AIE and followed by indomethacin after ChAT loss restored the ChAT level to that of rats never treated with ethanol, effectively reversing the AIE loss.2
The findings from the Morris water maze experiment aligned closely with Dr. Crews’ hypothesis. In the acquisition learning phase, the group of rats that received a placebo after undergoing the adolescent intermittent ethanol (AIE) exposure showed no difficulty in learning and completing spatial tasks, but showed deficits in the reversal learning task. In contrast, the group treated with indomethacin performed significantly better, similar to controls who were not treated with AIE.2
Overall, Dr. Crews concluded that the AIE model, which simulates binge drinking, significantly reduces cholinergic neurons through reversible mechanisms involving gene silencing and reversal of silencing mechanisms. He has added other studies supporting epigenetic mechanisms, such as increasing the expression of proinflammatory genes and silencing other genes in a reversible manner. These findings bring hope for treatments that may reverse the long-lasting effects of adolescent binge drinking and possibly also neurodegeneration.
1. Interview with Fulton Crews, P.h.D. 02/09/2024
2. Macht, Victoria, et al. “Indomethacin restores loss of hippocampal neurogenesis and cholinergic innervation and reduces innate immune expression and reversal learning deficits in adult male and female rats following adolescent ethanol exposure.” Alcohol: Clinical and Experimental Research, vol. 47, no. 3, 17 Feb. 2023, pp. 470–485, https://doi. org/10.1111/acer.15019.
3. “Fulton T. Crews, Ph.D..” Bowles Center for Alcohol Studies, 7 Feb. 2023, www.med.unc.edu/alcohol/faculty-research-2/ fulton-t-crews-ph-d/.

“If pregnant or breast-feeding, ask a health professional before use”: a common label many expecting mothers come across on almost all medicines, supplements, vitamins, foods, and drinks. There is no doubt that the diet of an expectant mother should be well balanced and carefully planned so that it consistently meets the needs of their own body, as well as their neonate’s. Not only are there a plethora of foods, drinks, vitamins, and supplements recommended for those expecting, but there are just as many suggested for them to avoid. Unsurprisingly, some of the most prominent substances expecting mothers are urged to avoid include drugs and alcohol
When a mother drinks alcohol during pregnancy, it reaches the unborn baby. Since the baby has no way to dispose of that alcohol, it keeps floating in the amniotic fluid which not only leads to miscarriage, fetal or infant death, and prematurity; but it also causes a range of birth defects (facial abnormalities and growth restriction) and neurodevelopmental disabilities (learning and cognition delays) collectively known as Fetal Alcohol Spectrum Disorder (FASD). Up to 1 in 20 US first graders may have
By Ria PatelFASD that not only impairs their physical health, behavior, and cognition, but also subjects them to a higher risk of metabolic disorders such as type II diabetes and cardiovascular disease at adolescence and adulthood due to exposure to alcohol before birth.1,2 In fact, extensive ongoing research is focused on minimizing the effects of FASD after the baby is born. However, studies investigating the effects of drinking alcohol on the mother and her metabolism – ones that relate maternal metabolism to fetus health – are unfortunately limited and rare.
Fortunately, Dr. Nipun Saini has acknowledged this issue, aiming her own research efforts on understanding how maternal and fetal metabolisms interact and influence one another in alcoholexposed pregnancies.1
Dr. Nipun Saini is an Assistant Professor of Nutrition at the Nutrition Research Institute (NRI) of the University of North Carolina at Chapel Hill. She is currently leading her own research projects to understand how the consumption of alcohol during pregnancy affects the metabolism of mothers and, therefore, the growth and development of their babies. Dr. Saini’s research interests have stemmed from a
history of enjoying learning about human biological sciences and a passion to make a clinical impact via research. Additionally, Dr. Saini’s impressive educational record and accomplishments have paved the way for her own success. She completed her PhD in biochemistry from the University of Nebraska-Lincoln in 2015, and recently ended her postdoctoral research atthe University of North Carolina at Chapel Hill as a K99 Awardee under the mentorship of Dr. Susan Smith. It was during her postdoctoral training that Dr. Saini got interested in research regarding alcohol and pregnancy. In fact, nohhhht only did her postdoctoral work influence her interest in pregnancy metabolism, but, during that time, she herself was an expecting mother – her work effectively became personal!

During her postdoctoral research with Dr. Smith, Dr. Saini’s initial project investigated the impact of alcohol on pregnancy and iron metabolism in the mother and her fetus. Their findings suggested that alcohol causes inflammation in the mother and fetus. This was accompanied by fetal anemia, an accumulation of iron in the fetal liver and decrease in brain iron content. In consequence, the inadequacy of brain iron content of the fetus causes a plethora of neurodevelopment deficits and cognition deficits.3 Importantly, when mothers were provided with iron fortified diets, alcohol’s impact on maternal inflammation, fetal anemia, and brain iron content was reversed.5
While learning from her research, and currently being an expectant mother herself at the time, Dr. Saini quickly noted the sheer importance of needing proper nutrition and a balanced diet during pregnancy – so much so that the slight deficiency of any macro- or micronutrient (like iron) can interact with alcohol, and the consequences can be dire. Thus, Dr. Saini wanted to continue to understand how alcohol contributes to fetal growth impairments and neurodevelopmental deficits because of a mother’s nutrient and alcohol intake choices. So, she began her own research project as a postdoc looking at how glucose metabolism is altered in mothers when they drink alcohol during their pregnancy and is continuing this research as an Assistant Professor at the University of North Carolina at Chapel Hill NRI.

Currently, Dr. Saini is looking into where the glucose is going in alcoholexposed pregnancies, especially since it is not reaching the fetus or going into muscle stores, as evidenced by looking at other bodily tissues. Along with limited glucose, Dr. Saini’s research also suggested that pregnant drinkers have a fatty liver due to an increase in fatty acid synthesis. She is also in the process of understanding the mechanism behind this fatty liver accumulation and whether the glucose, as from her first point of curiosity, is getting converted to fat and stored as fatty liver tissue.
The consequences of the consumption of alcohol during pregnancy are not unknown
In her most recent scientific publication, Dr. Saini has addressed that when mothers drink alcohol during their pregnancy, their glucose levels decrease as their livers stop producing additional glucose.5 These mothers also become insulin sensitive. This is especially detrimental during the third trimester, when most of the metabolic adaptations occur in a mother to ensure that she spares her glucose for her fetus, which uses it for its energy.1,5 This lack of maternal glucose production and supply then affects the fetal brain by decreasing its source of energy and causing a consistent 5% decrease in fetal weight.5
Dr. Saini’s most recent research endeavors have been conducted in mouse models. Although the effects of alcohol in her mouse models cannot be directly translated those in humans, this project has been an incredible steppingstone for Dr. Saini to drill down into the maternal metabolism mechanisms that change or result from alcohol consumption.
Clearly, Dr. Saini’s current efforts serve to bridge the gap in knowledge concerning FASD and changes in fetal health as a result of alcohol-exposed pregnancies via maternal metabolism mechanisms. She eventually hopes to create and implement targeted, streamlined interventions for mothers who have been exposed to alcohol during their pregnancy so the long-term effects on their fetuses can be mitigated. Targeting issues like FASD at the root—in other words, starting with the mother—
is encouraged as a more precise and preventative form of clinical intervention. Thanks to Dr. Saini, we are now one step closer in understanding how alcohol contributes to fetal health complications because of maternal nutritional deficits and altered maternal metabolism!
1. Interview with Nipun Saini, Ph.D. 01/23/2024
2. Alcohol use in pregnancy. https:// www.cdc.gov/ncbddd/fasd/alcohol-use. html (accessed February 2nd, 2024).
3. Huebner, S. M.; Blohowiak, S. E.; Kling, P. J.; Smith, S. M. The Journal of Nutrition 2016, 146, 1180–1188.
4. Saini, N.; Helfrich, K. K.; Kwan, S. T. C.; Huebner, S. M.; Abazi, J.; Flentke, G. R.; Blohowiak, S. E.; Kling, P. J.; Smith, S. M. Alcoholism: Clinical & Experimental Research 2019, 43, 2332–2343.
5. Saini, N.; Mooney, S. M.; Smith, S. M. The FASEB Journal 2023, 37.

By now, the term “quarantine baby” often gets thrown around in conversation as a humorous reference to the unique social development of children raised during the pandemic. Scientists and the general public share the assumption that the COVID-19 pandemic and the isolating nature of subsequent lockdowns affected psychological well-being, especially for young children, society’s most impressionable members.
Dr. Margaret Sheridan is an associate professor in the Clinical Psychology department at the University of North Carolina at Chapel Hill and the director of the Child Imaging Research
on Life Experience (CIRCLE) Lab. As the principal investigator of this lab, Dr. Sheridan leads various studies that examine the neural mechanisms through which adversity influences brain development and psychopathology, specifically in early childhood. An ongoing initiative from this lab is the Wellness Health and Life Expectancy (WHALE) study; this study uses a combination of emotional regulation challenges and physiological measures to assess brain development and psychopathology risk in children aged 4 to 7 years old. This data collection effort was active when the pandemic unexpectedly swept the country. The chaos and uncertainty of the

pandemic created a unique social context that proved valuable for social and developmental psychological research. The initial weeks of the pandemic and the resulting stay-at-home order created widespread confusion, leaving families grasping at a semblance of structure. Dr. Sheridan remembers feeling acutely concerned about the families in her research group. She recalls, “I’m having such a hard time regulating myself and regulating my kids, the whole world had become so anxious all of a sudden.” This was a stressful time for anybody but particularly for the at-risk families in the WHALE sample. These children and their families were already coping with low resources and high risk for exposure to family violence; these adverse factors were amplified by the pandemic. As a means of documentation and support, Dr. Sheridan’s lab conducted weekly surveys to assess the effects of COVID-19 related disruptors on psychological well being. These self-reported surveys included items that measured family violence exposure, caregiver anxiety, child anxiety, and presence of disruptors.²
Dr. Sheridan and her coauthors found intriguing patterns related to family violence throughout the 15-month period. They observed an initial spike in the family violence score over the first four months of the pandemic; this score is constructed from measures of corporal punishment and partner violence.2 After this initial increase in rates of observed violence, the scores did not remain consistently high; they decreased dramatically. Dr. Sheridan
interprets this pattern as a reaction to the initial unpredictability of COVID-19. The sudden increase in family violence can be attributed to the pandemic acting as a major disruptor for family routine and support systems, but this pattern stabilized when these family units received more predictability. For example, when the CDC released guidelines regarding COVID protocol or when the first vaccine was approved by FDA; these events likely provided families with hope and increased feelings of security. This is critical information for policymakers and government officials to consider because understanding the factors that contribute to family adversity and the structures that eventually lower the frequency of such behavior can inform policies that mitigate rates of family violence (Figure 1).
Considering this pattern, Dr. Sheridan was inclined to collect data on anxiety and depression because family violence is associated with psychopathology risk in kids. Similar to rates of family violence, there was an initial spike in anxiety characteristic of the uncertainty associated with early pandemic life. However, unlike rates of family violence, the pattern in anxiety fluctuated frequently over time. As the pandemic progressed, the researchers observed a fascinating pattern of anxiety rates between family members. Parent anxiety patterns tracked children’s anxiety very closely, illustrating a relationship where caregiver anxiety patterns predict their children’s subsequent anxiety.2 This predictive relationship is conducted through various caregiver pathways including amplifying threat perception in children, overcontrolling parenting styles that prevent children from developing coping mechanisms, and reinforcing anxious processing biases.1
In contrast, depression had a distinctly different pattern. Unlike anxiety,

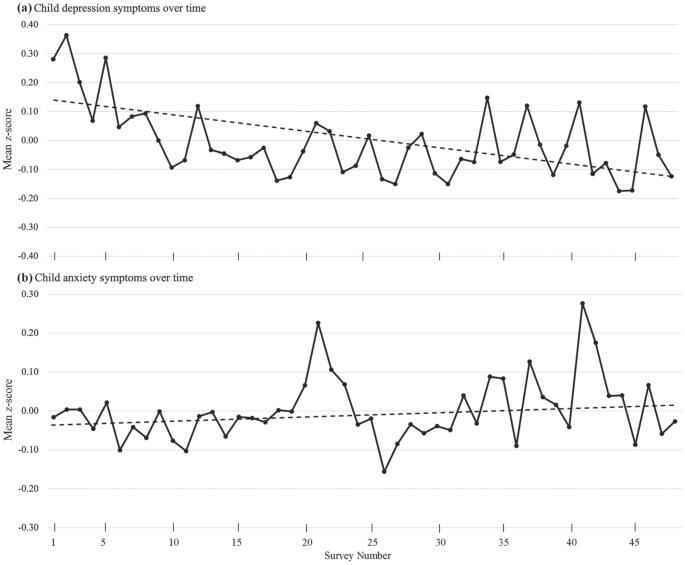
a factor that depicted variation across time, depression in children increased steadily over the duration of the pandemic. This distinction between anxiety and depression patterns contributes to a significant ongoing conversation among clinical psychology academics regarding co-occurrence of symptoms, a debate examining if this correlation is the result of a causal relationship between anxiety and depression or if it’s an indication that these separate diagnoses are the same phenomenon. Dr. Sheridan’s data on the distinct patterns of anxiety and depression over the course of the pandemic contributes to the argument that these are entirely separate disorders functioning independently of each other (Figure 2).
Dr. Sheridan hopes to expand on this research by extracting specific connections between spikes in psychopathology and related contextual events. In many ways, the pandemic acted as a naturally-occurring experiment that many researchers harnessed to produce important documentation. Ironically, this proved to be a challenging time to maintain faith in science. Dr. Sheridan remarked, “staying convinced that science was a way to solve problems in that moment was hard.”
Studying COVID-19 was an academic coping method, a way of focusing on
something controllable while the world had suddenly become so uncertain. The choice to continue research efforts during the pandemic produced valuable data. This work revealed the pattern of family violence during the pandemic, the predictive relationship between caregiver anxiety and subsequent child anxiety and depicted the distinctly separate models of anxiety and depression. Moving forward, Dr. Sheridan is interested in observing big markers of change across this pandemic period to specifically correlate major events to spikes in psychological wellbeing. This includes societal events like the Black Lives Matter protests, COVID-19 related disruptors, and political unrest. Linking these societal events with family experiences could reveal significant details about the influence of contextual factors on the well-being of family units. Furthermore, this data has potential to inform important public policy related to family and child wellbeing.
1. Interview with Margaret Sheridan, Ph.D. 02/13/2024.
2. Gruhn, M.; Miller, A.B.; Machlin, L.; Motton, S.; Thinzar, C.E.; Sheridan, M.A. Res Child Adolesc Psychopathol 2023, 51, 233–246.

Have you ever questioned whether you are learning and studying in the best possible way? Dr. Mulligan and his research team are studying the testing effect, the finding that retrieving information from memory is more beneficial than alternative learning experiences, like restudying or rereading.
Dr. Neil Mulligan received his bachelor’s degree in computer science and psychology from Duke University. He continued his education at the University of North Carolina at Chapel Hill, receiving his Ph.D. in cognitive psychology in 1994. Additionally, he has been serving as a professor in the psychology department at UNC since 2002.1
In graduate school, Dr. Mulligan was broadly interested in cognitive psychology and, through his advisor, began to research higher cognitive functions, such as memory.1 Since then, his extensive background in memory has enabled him to ask and answer more complex questions concerning the subject.
“Until recently, the research I did was very much basic research on topics of memory, learning, and attention. But a couple of the graduate students I have had recently were very interested in application to educational settings.”1
“The high-end goal of education is to create experts.”
Dr. Mulligan and his team have been investigating the testing effect. This kind of information retrieval typically takes place in the form of a test. Introducing the findings from testing effect studies could impact the way students learn. However, further research must be conducted to ensure the testing effect applies to generally to many types of students across many settings. Dr. Mulligan and his team investigated the impact of prior knowledge on the testing effect.1
Dr. Mulligan and Zachary Buchin, one of his previous graduate students, experimentally manipulated participants’ prior knowledge to study the effect on the learning benefits of retrieval. The manipulation of prior knowledge is crucial to generate a causal relationship because individual differences such as general cognitive abilities, interests, or motivations can create limitations without this manipulation.1
A total of 128 participants’ prior knowledge on subjects of either geology or sensation and perception was experimentally manipulated over 3 days through training phases. Then, they were divided into two conditions—restudy or retrieval practice (testing). Participants were either given 8 key ideas and examples to restudy or 8 practice questions with feedback for each topic
Figure 1. Overview of the study procedure for the training, learning, and testing phases in Retrieval-Based Learning and Prior Knowledge study. Figure courtesy of Buchin et al.
within their learning subject. Finally, two days later, they took a final test to assess topic mastery.2
The study revealed that, on average, participants that were subject to retrieval practice performed better on the final test. More importantly, there was no evidence to suggest that retrieval practice affected those with high (HPK) or low prior knowledge (LPK) differently, suggesting that the testing effect enhanced learning for all subjects, whether they had substantial prior knowledge about this topic or not.2
Whether it is an introductory or a high-level course, these findings imply that professors should not have to worry about

knowledge topics; LPK = low prior knowledge topics). Figure courtesy of
levels of prior knowledge when utilizing the testing effect. Dr. Mulligan emphasized the importance of study replication and the need to research more individual differences and other factors to fully understand the extent of the testing effect.1
Furthermore, other research is being conducted in the cognitive community to examine how other individual differences may affect the testing effect. In addition to Dr. Mulligan’s prior knowledge studies, his team has been researching if the testing effect works not just for fact recall but also for transitive inferences—drawing conclusions and creating complex knowledge structures.
According to Dr. Mulligan, the high-end goal of education is to create experts, and that cannot be done solely through fact recall. “Expertise is situations where people have learned many different facts, and they have synthesized and combined those facts into complicated knowledge structures. Then they can apply [those structures] to novel situations in their domains of expertise.”1 However, another study they conducted found that learning through the testing effect method can make it harder to create complicated knowledge structures and draw
transitive inferences.2
Dr. Mulligan stated that this was a surprising finding and that they need to further replicate and investigate this study in less lab-controlled environments and more real-life educational experiences to draw more conclusions about the testing effect.1 Dr. Mulligan stressed the importance of replicating studies and continuing to investigate other individual and situational differences to fully understand the limitations of the testing effect. His research will continue to help educators and students incorporate the best ways to learn and study.

Dr. Neil Mulligan
1. Interview with Neil Mulligan, PhD. 02/15/2024
2. Buchin, Z. L. (2022). Retrieval-based learning and prior knowledge. Journal of Educational Psychology, 115(1), 22. https://doi-org.libproxy.lib.unc.edu/10.1037/edu0000773
3. Mulligan, N. W., Buchin, Z. L., & Powers, A. (2023). Transitive inference and the testing effect: Retrieval practice impairs transitive inference. Quarterly Journal of Experimental Psychology, 76(10), 2356-2370. https://doi-org. libproxy.lib.unc.edu/10.1177/17470218231156732

Metabolism is the process of breaking down food into energy and has many helpers for doing so.
Fats, scientifically known as lipids, are one of three sources that humans naturally use for energy. Uncovering how hedgehog proteins—which are native to humans—and vital for molecule signaling like C. elegans can provide a great deal of valuable insight into their use of lipids.3 Learning more about reasons behind having a healthy diet and exercise is important for nurturing the resource that gives the energy to accomplish so much every day. The goal of Dr. Dowen’s research team is to dive deeper into how organisms best use lipids to accomplish certain tasks.
Dr. Dowen’s research achievements extend from a vast background of expertise, from a bachelors in science in biology at the University of North Carolina at Chapel Hill to completing a PhD at the University of San Diego.1 Additionally, he refined his research skills through achieving a post doctorate fellowship at Massachusetts General Hospital/Harvard and upon doing so, started his own research that focuses on fat metabolism. The goal of his research is to understand the makeup, storage process, and use of fat during the body’s metabolic process.1 By studying hedgehog proteins and organisms like C. elegans roundworms that regulate fat metabolism similarly to humans, an easier connection can be made for preventing metabolic disease and promote a stronger reason for staying healthy in the process.

In the human body, fats are managed through communication between tissues via secreted proteins. The line of communication decides whether lipids are stored for
later use or released for energy needs. Hence, there are two big questions that Dr. Dowen’s lab tries to answer: how the human body controls fat levels as it grows, and whether this path of communication involves team players other than proteins.2 Hedgehog proteins are well known for regulating human development but there’s still must discover if they also play a role in metabolism. C. elegans nematodes and hedgehog proteins within the body’s intestines are used currently in the lab to better

Figure 1. A) PQM-1 working with other proteins to stop certain genes from turning on. B) Shows adult animal genes being turned on after receiving different treatment during experimentation. C) Shows PQM-1 within the intestine of young and adult animals. D) Adult animal genes being turned on during metabolism after exposed to different treatments. Image courtesy of Dr Dowen.

Figure 2. A and B) Researchers using microscope and genetic experiments to study how miRNA affects fat release from the intestine. C) Microscopic view to see if different gene mutations affect fat related or adult genes. D) Conducted genetic tests on mutant animals to see how different genes affect fat genes. E) A diagram that’s showing when genes become active and what they target during larvae stage. F) Shows how silenced genes affect fat genes of certain animals. Image courtesy of Dr Dowen.
understand how fat development is managed.
Beyond their general significance, both C. elegans and hedgehog proteins can offer insights into defining how fat metabolism works. C. elegans are tiny roundworms capable of storing fat in their bodies like humans; hedgehog proteins are responsible for how human cells grow and organize body tissue. Dr. Dowen described his first experiment concerning the process in which C. elegans control how fat moves. In mature animals, C. elegans contain fats that are normally stored and transferred from the intestine to their reproductive cells for their offspring. To determine how this is possible, Dr. Dowen experimented with C. elegans roundworms that display a green luminous color when fat regulation starts, allowing for fat production tracing. Specific genes in the worms were silenced, or turned off, in order to identify major players in fat regulation, which were found to be tiny molecules called miRNAs (which control fat production in the worm’s intestine). The lab also discovered other key players in fat metabolism. For example, the hormone insulin plays a large part in how diabetes arises while controlling fat production in the C. elegans intestine. Moreover, the protein SGK-1 directs when and where fat production can occur, as it slows down fat production by the metabolic regulator PQM-1. Furthermore, Dr. Dowen’s research on C. elegans explores how stress can be another factor that affects fat metabolism reproduction.4 Two important molecules he experimentally discovered were CEH-60/PBX and UNC-62/MEIS that are transcription factors used to control how genes are expressed. They work together along with PQM-1 to control fat metabolism movement from mother to offspring for a smooth transition.3 The molecules also play a larger role in the lifespan of C. elegans
worms and how well the organisms respond to stress. Dr. Dowen came to the realization that if these key players were to undergo any issues, the malfunction would cause an improper storage of fat, extend the life span of the worms, and bring about higher resistance to stress.
Beyond research endeavors with studying fat metabolism, Dr. Dowen and his lab are all driven by a passion for medical progression. The lab’s biggest goal is to address human pathways that commonly go awry associated with metabolic disorders— such as cancers, diabetes, and high blood pressure—while also promoting an agenda for healthy living habits. The main objectives of the research are discovering if new molecules are involved in the fat metabolism pathway, how the interaction works when proteins and tissues communicate with one another, and connecting the dots with how one protein talks to another.2 Ultimately, cracking the lipid code, can create a new guide for self-care and save more lives in the process.

Figure 3. A) Showing experimentation to see how mTORC2 affects the cell development of worm cells. The research team looked at images of worms when their genes were turned on. B) Show different parts of mcTORC1 and mTORC2 (proteins that are important for cell development) that are compared to proteins of humans and C. elegans roundworms. C) Images of worm growth under different conditions to see how it would affect cell development. D) Reveals observations of turned off genes affecting cell activity of other genes in worm cell development using a technique called RT-qPCR.
1. Dr. Rob Dowen, Ph.D
2. Interview with Rob H. Dowen, Ph.D. 02/01/2024.
3. Dowen, R. H.; Breen, P. C.; Tullius, T.; Conery, A. L.; Ruvkun, G. A MicroRNA Program in the C. Elegans Hypodermis Couples to Intestinal MTORC2/ PQM-1 Signaling to Modulate Fat Transport. Genes & Development 2016, 30 (13), 1515–1528.
4. Dowen, R. H. CEH-60/PBX and UNC-62/MEIS Coordinate a Metabolic Switch That Supports Reproduction in C. Elegans. Developmental Cell 2019, 49 (2), 235-250.

n the dynamic ecosystem of our intestines, a multitude of bacteria collaborates to orchestrate functions essential for our well-being. However, when this delicate balance is disrupted, chaos ensues.1 Dysbiosis, a deleterious imbalance in gut bacteria composition, frequently emerges in inflammatory bowel diseases (IBDs) like Crohn’s and ulcerative colitis, affecting approximately 3.1 million adults in the United States alone.2 Under these conditions, the intestines often become home to heightened colonization of E. coli and subsequent reduced microbial diversity. Specifically, a strain known as adherent invasive E. coli (AIEC) is believed to contribute to the ongoing inflammatory activity in these diseases.3
Unlike pathogenic strains of E. coli, AIEC do not produce the typical toxins that cause gastrointestinal distress. Rather, they possess the ability to adhere to and infiltrate epithelial cells and survive in macrophages—a quality believed to contribute to the persistent in-vivo inflammation of inflammatory bowel disease.4
While considerable research has been devoted to these bacteria, the descriptive AIEC definition is based primarily on in-vitro cell experiments, raising uncertainty about its true predictive power regarding their behavior in the human body. However, assessing the abundance of AIEC in patients is challenging as they may be genetically similar to other E. coli strains while functionally distinct, impeding differentiation of strains residing in the gut. This inherent similarity is an obstacle prevents researchers from truly understanding how AIEC can influence the composition of our microbial community and interactions with the host, including the molecular features that enable it to colonize inflamed mucosa. Obtaining these insights is crucial in identifying inflammatory bowel disease patients that may harbor high-risk E. coli strains, thus informing targeted interventions that go into greater detail than the traditional in vitro classification.4
Dr. Janelle Arthur, a microbiologist and immunologist at The University of North Carolina at Chapel Hill, endeavors to address the intricacies of identifying and distinguishing E.
coli in inflammatory bowel diseases. Devising a more comprehensive approach to identify E. coli strains capable of intimately colonizing the mucosa and inducing disease, her research stands to revolutionize our understanding of gut microbiota interactions by reclassifying bacteria based on their phenotypes and function.4

The exploration of the microbiome and its dynamic interplay has often been limited to techniques like stool sampling and traditional microbiome sequencing, which offers insights primarily into our fecal microbiota. However, “[one] also has a community of bacteria that is adherent to the epithelium and in the mucus layer that protects [one’s] cells, and [one] doesn’t capture that when [one] gets a fecal sample and does microbiome sequencing” as Dr. Arthur explains. To elucidate the
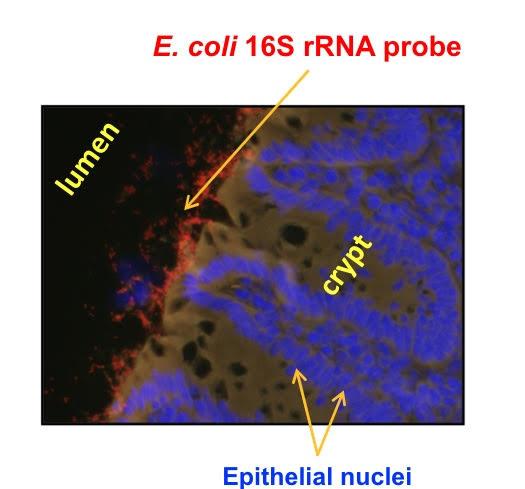
2. E. coli (red) in a cross section of a colon. Image courtesy of Dr. Janelle Arthur.
complexities surrounding the ability of identifying which E. coli can actually intimately colonize the mucosa and trigger inflammation, Dr. Arthur and her colleagues have explored and employed a technique known as molecular barcoding.4
To implement molecular barcoding, strains of non-AIEC or AIEC isolated from humans were marked with an antibiotic resistance gene and a molecular barcode was inserted into a neutral chromosome region.This meticulous process ensures each strain can be uniquely identified in the microbial environment since there is no single, common genetic feature present across all strains. By incorporating an antibiotic resistance gene, the researchers introduce another genetic marker that facilitates the tracking and identification of individual bacterial strains within complex microbial communities. Simultaneously, inserting an identifiable molecular barcode into the chromosome region further enhances the specificity and precision of identification, allowing for the accurate discrimination of E. coli strains and quantification. The next crucial step entailed administering fecal transplants into mice models to colonize them with gut bacteria, thereby simulating a microbial environment. By introducing these bacterial communities into the mice, the researchers created a dynamic and physiologically relevant system for studying the interactions between the gut microbiota and the host immune system, as well as many other possibilities. Utilizing the efficiency and versatility of this technique offers significant advantages over studying the microbiome as it allows for rapid and comprehensive analysis of bacterial diversity and dynamics in an in-vivo system. As Dr. Arthur notes, “We know that our microbiome adapts to us over time, so if we can always tag that strain because it has the barcode, we can look over time with whole genome sequencing and see how it changes over time in

response to different triggers…”.4
With molecular barcoding, both AIEC and non-AIEC strains demonstrated consistent colonization of the mouse intestines. Additionally, during inflammation, both strain types demonstrated heightened abundance in inflammation susceptible mice, effectively displacing other bacteria and altering the microbiota composition. Upon further examination, distinctive genomic traits were revealed in four highly colonizing E. coli strains—two AIEC and two non-AIEC varieties. These genetic features provided the bacteria with metabolic advantages, such as iron acquisition and carbohydrate utilization. Though more studies are necessary, these findings both highlight the intricate dynamics of E. coli within the host environment and challenge the conventional qualitative definition of AIEC, calling for a reevaluation of its relevance in vivo.3

4. Rachel Bleich and Chuang Li. Image courtesy of Dr. Janelle Arthur.
Navigating these findings was no small feat, as the study demanded a highly interdisciplinary approach and a wealth of expertise. Dr. Arthur collaborated closely with two postdoctoral students, Rachel Bleich and Chuang Li, whose diverse skill sets provided instrumental support. Bleich’s proficiency in animal inflammation biology and microbe-microbe interactions facilitated the execution of the mouse models while Li’s expertise in bioinformatics translated complex data into comprehensible insights, seamlessly integrating knowledge into a comprehensive synthesis of their findings. Dr. Arthur’s research marks a significant forward progress in our understanding of inflammatory bowel diseases and their treatment strategies. By establishing a robust method for studying a wide range of E. coli strains and identifying associated features of mucosal colonization through molecular barcoding, this research offers invaluable insights not only into the true mechanisms underlying IBD, but also lays the foundation for future discoveries that can improve outcomes and quality of life for IBD patients worldwide. 4
1. Ogunrinola, G. A., Oyewale, J. O., Oshamika, O. O., & Olasehinde, G. I. (2020). The Human Microbiome and Its Impacts on Health. International Journal of Microbiology, 2020, 1–7. https://doi.org/10.1155/2020/8045646
2. CDC. (2020, May 21). People with IBD Have More Chronic Diseases. Www.cdc.gov. https://www.cdc.gov/ibd/features/ IBD-more-chronic-diseases.html
3. Bleich, R. M., Li, C., Sun, S., Ahn, J.-H., Dogan, B., Barlogio, C. J., Broberg, C. A., Franks, A. R., Bulik-Sullivan, E., Carroll, I. M., Simpson, K. W., Fodor, A. A., & Arthur, J. C. (2023). A consortia of clinical E. coli strains with distinct in vitro adherent/invasive properties establish their own co-colonization niche and shape the intestinal microbiota in inflammation-susceptible mice. Microbiome, 11(1). https:// doi.org/10.1186/s40168-023-01710-y
4. Interview with Dr. Janelle C. Arthur, PhD., 02/07/2024

In the depths of the intestines lies a world of microscopic cells with immense power over one’s health. Meet the enteroendocrine cells: Though small in number, these specialized cells have significant influence over metabolism and overall well-being. Scattered along the digestive tract, enteroendocrine cells hold the key to regulating nutrient absorption, hunger, and even insulin secretion.1 Their malfunction has been linked to a plethora of medical conditions, from inflammatory bowel disease to obesity and diabetes.2 By exploring the mechanisms of enteroendocrine cells, new avenues for combating these pervasive health challenges can be discovered.
Enter Dr. Heather A. McCauley, a trailblazer in the field of gastrointestinal research whose work is currently based at the University of North Carolina at Chapel Hill.2 She completed her undergraduate studies at the University of Southern California, where her experience in a lab studying fatty acid metabolism in skeletal muscle sparked a passion for bench research, leading her to pursue a career of scientific discovery. After studying the TGFbeta signaling pathway in both cancer and eye differentiation during her Ph.D. program in Molecular and Developmental Biology from the University of Cincinnati, Dr. McCauley sought a field in research aligned with her personal interest in nutrition and metabolism.2 Dr. McCauley was intrigued by the oftenoverlooked role of intestinal metabolism, which accounts for significant oxygen consumption and serves as the primary site for nutrient sensing and absorption.2 In particular, she was fascinated by enteroendocrine cells, which comprise only

Figure 1. Depiction of how the PYY may regulate ion, water, and nutrient transport in the small intestine in the presence and absence of entroendocrine cells (EECs). Figure courtesy of McCauley et al.
one percent of the intestinal epithelium but secrete about 20 different hormones crucial for systemic regulation.1 Contrary to previous assumptions, these cells play vital local roles in supporting nutrient absorption and maintaining intestinal stem cell equilibrium, thus guiding Dr. McCauley’s current research pursuits in unraveling the complexities of gut biology.2

Dr. Heather McCauley
Dr. McCauley’s research delves into the intricate world of enteroendocrine cells, uncovering their role in shaping the gut’s physiology.2 In regards to Dr. McCauley’s methodology, her lab combines mouse models and human pluripotent stem cell-derived organoids, which are simplified models of organs. Acknowledging the necessity for human relevance, she strategically navigates between the complexities of mouse physiology and the simplicity of cell cultures. Using this dual-model approach, Dr. McCauley studies cellular mechanisms in vitro and verifies their physiological implications in vivo.2
Her investigations focus on mice deficient in the Neurogenin-3 gene, which is vital for enteroendocrine cell development.2 Utilizing the villin promoter—a DNA sequence that regulates gene expression of the villin protein, which is produced in intestinal epithelial cells responsible for nutrient uptake—results in targeted deletion of Neurogenin-3. This deletion leads to pronounced malabsorptive symptoms in embryonic mice, including severe lack of weight and diarrhea.3 However, confronted with the challenge of early mortality in these models, Dr. McCauley employs an innovative solution: a tamoxifen-inducible villin-cre-ER system. The system allows for deletion of Neurogenin-3 post-intestinal development, enabling the exploration of enteroendocrine cell effects without the confounding influence of lifelong malabsorption.2 This meticulous approach unveils the acute impacts of enteroendocrine cells on intestinal function, contributing vital insights to the field of metabolic research.
Notably, a rare subset of children is born with mutations in Neurogenin-3, resulting in the absence of enteroendocrine cells
in their intestines. This results in severe malabsorptive diarrhea akin to the phenotype observed in mice lacking these cells. Patients often require intravenous nutrition supplementation in order to survive, with some undergoing small bowel transplants due to the condition’s severity.3 Dr. McCauley’s research unveiled a potential breakthrough when studying mice lacking enteroendocrine cells from birth: administration of peptide YY (PYY), a hormone produced by the cells, rescued the mice from premature death. Daily supplementation with PYY led to normalization of intestinal electrophysiology, facilitating nutrient absorption and leading to weight gain and improved survival rates. Dr. McCauley explored the prospect of translating these findings into clinical practice by collaborating with a pediatric surgeon specializing in disorders involving infants who do not absorb enough nutrients.2 While the status of clinical trials remains uncertain due to personnel changes, the promising outcomes in this preclinical model underscore the therapeutic benefits of PYY supplementation in addressing nutrient absorption disorders.
transmit crucial information throughout the body. This unlocks a new understanding of how the gut epithelium maintains its integrity and regenerates over time.
“Dr. McCauley seeks a balance between simplicity and mechanistic insights”
Reflecting on the biggest discoveries of the McCauley Lab, Dr. McCauley emphasized two primary findings. The first discovery was that enteroendocrine cells can regulate the electrophysiology of their neighbors.2 Nutrients are absorbed through association with ions such as sodium.4 If the gradient across that apical membrane is disrupted, then absorption won’t occur properly. The McCauley Lab found that enteroendocrine cells regulate this process.2 This finding depicts the intricate interplay between different cell types within the gut, not only shedding light on the mechanisms behind nutrient absorption but also on potential targeted therapies in conditions where nutrient uptake is compromised.
The second discovery was that enteroendocrine cells can regulate intestinal stem cell activity, which is responsible for maintaining the epithelium during homeostasis and regeneration. Essentially, intestinal stem cells can sense nutrients and change how much they proliferate.2 If mice are on a high fat diet or mice are fasted, their intestinal stem cells will change.5 Enteroendocrine cells tell intestinal stem cells whether nutrients are present or not, acting as an intermediary to

In addition to these primary findings, the McCauley Lab’s research sheds light on the dysregulation of enteroendocrine hormone expression, linking it to various diseases such as inflammatory bowel disease (IBD), obesity, and Type II diabetes. For instance, in IBD, there is an increase in enteroendocrine cells accompanied by alterations in hormone production, whereas conditions such as obesity are associated with reduced enteroendocrine cell populations.2 Dr. McCauley’s work in this area extends beyond understanding disease mechanisms; it also informs therapeutic interventions. Drugs targeting enteroendocrine hormones—such as Ozempic and Wegovy—capitalize on the fact that some enteroendocrine hormones act on the pancreas to increase insulin secretion and improve glycemic control in individuals with Type 2 diabetes.2 This understanding of enteroendocrine cell function not only deepens the knowledge surrounding disease pathogenesis but also opens up promising avenues for treatments.
While the McCauley Lab has made significant strides in their research, they face a challenge in reconciling findings between tissue culture and mouse models. It is challenging to distinguish between essential local intestinal processes and complex systemic influences. To address this, they must decipher the physiological interactions in mice and identify crucial components within tissue culture systems, such as the roles of immune cells and enteric neurons in observing phenotypic changes.2 Dr. McCauley seeks a balance between simplicity and mechanistic insights, aiming to optimize model systems for high throughput experimentation while ensuring biological relevance.
Looking towards the future of her research, Dr. McCauley finds the most promise in answering the question, “If we alter [one’s] diet and include supplements, can [one] then produce more of the hormones that regulate insulin secretion and regulate hunger and satiety to ultimately improve the ability to absorb nutrients?” The approach presents a promising avenue for personalized medicine tailored to individual gut health. By furthering the understanding of enteroendocrine cell function, Dr. McCauley charts a course towards a future where “the common person knows that they can alter their gastrointestinal physiology by ingesting something different.”2
1. McCauley, H.A. J. Nutr. 2020, 150, 10-21.
2. Interview with Heather A. McCauley, Ph.D. 2/16/2024
3. Zhang, X.; McGrath, P.S.; Salomone, J.; Rahal, M.; McCauley, H.A.; Schweitzer, J.; Kovall, R.; Gebelein, B.; Wells, J.M. Dev. Cell 2019, 50, 367-380.
4. McCauley, H.A.; Matthis, A.L.; Enriquez, J.R.; Nichol, J.T.; Sanchez, J.G.; Stone, W.J.; Sundaram, N.; Helmrath, M.A.; Montrose, M.H., Aihara, E. et al. Nat. Comm. 2020, 11, 1-10.
5. Enriquez, J.R.; McCauley, H.A.; Zhang, K.X.; Sanchez, J.G.; Kalin, G.T.; Lang, R.A.; Wells, J.M. Cell Rep 2022, 41, 1-14.
Imagine throwing a wrench into an engine running at breakneck speed—the engine would explode and suffer huge damage! What if the engine was barely moving? Then, the engine may cease to run, but it would not suffer much damage. If the wrench were removed, it may even resume normal function. This is what happens on a molecular level when a pathogen is tolerant to antibiotics. Antibiotics typically kill proliferative, metabolically active bacteria effectively, but are unsuccessful when it comes to slow-proliferating, metabolically inactive bacteria. They function by disrupting cell processes within the cell after target binding, ultimately resulting in cell death. In antibiotic resistance, the antibiotic fails because bacteria find a way to stop target binding, perform the process blocked by the antibiotic, or break down the antibiotic itself. On the other hand, pathogens do not defend themselves against antibiotics in antibiotic tolerance. Instead, the downstream effects of the antibiotics’ target binding are not lethal because the cell is in an energy-poor state.
The subpopulation of bacteria that are in this energy-poor state and persevere through antibiotic treatment are known as persisters. Persister cells of pathogens cannot grow and proliferate in the presence of antibiotics. However, they survive until the antibiotic is removed, at which point they can regrow. The Conlon Lab at the University of North Carolina at Chapel Hill researches antibiotic tolerance with two main goals: examining the role of the human body’s immune response on persister formation and improving antibiotic efficacy by discovering methods to combat persisters.
Dr. Brian P. Conlon joined the University of North Carolina at Chapel Hill as an Assistant Professor in August 2016, and became an Associate Professor in 2021, investigating the clinical relevance of antibiotic tolerance. His work specifically focuses on the Staphylococcus aureus species, a major bacterial human pathogen that is the leading cause of skin and soft

tissue infections. He obtained a PhD at the University College Dublin in Ireland exploring biofilm formation in Staphylococcus epidermidis and its association with higher treatment difficulty.1 Biofilm is a protective shield formed by bacteria that blocks treatment from killing an infection.2 Dr. Conlon became interested in how biofilms withstood antibiotics, leading him to pursue his postdoctorate with Kim Lewis, an expert in persister formation, at Northeastern University. Under Lewis’ mentorship, Dr. Conlon discovered that “energy levels were a major predictor of bacterial survival to antibiotic challenge and that persister cells were a subpopulation of cells with a really low level of ATP, the energy currency of the cell.”1 He also identified a class of drugs able to target bacteria without depending on ATP levels: those whose target was to activate a protease (an enzyme that breaks down protein) and cause the cell to self-digest. After his postdoctorate, Dr. Conlon wanted to take his research to the next level: the human body.
Dr. Conlon states that “we have a fairly good understanding of how antibiotics work against bacteria in tubes, but comparatively very poor understanding of what they’re actually doing in vivo”.1 Antibiotics must be thought of in the context of the body’s immune response. An important finding in Dr. Conlon’s research is that reactive oxygen species produced by phagocytes can push S. aureus into a metabolically dormant state in vivo (during infection in a living organism). Phagocytes are a type of immune cell that surround and kill foreign microorganisms. S. aureus is a pathogen that can live and grow in macrophages (white blood cells that are phagocytes)
after being taken up. When it fails to kill S. aureus, the phagocyte pushes the S. aureus into a metabolically dormant phase so that the bacteria does not harm the host. This then reduces antibiotics’ ability to kill the pathogen. Human immune systems did not evolve in the presence of antibiotics, so their innate response to infections sometimes work in opposition to how antibiotics are meant to treat infections. This is troublesome as we rely on antibiotics to be most effective when the host is not able to clear infections themselves.

Figure 1. Methicillin-resistant Satphylococcus aureus (MRSA). Figure courtesy of NIAID/NIH vis Wikimedia Commons.
The antagonism between the innate immune response and our antibiotics is most likely the root cause of long-term antibiotic treatment in the clinic and contributes to high treatment failure and mortality rates.
A key defense mechanism of the immune system is respiratory burst: the rapid release of reactive oxygen species for pathogen killing. In the study, when macrophages were stimulated into an inflammatory state, respiratory bursts would produce reactive oxygen species and reactive nitrogen species that induce S. aureus into a metabolically dormant state.3 The status of macrophages were observed using fluorescent antibodies and reactive oxygen and nitrogen species levels were measured using different dyes and markers. Mouse models were used to test the effect of certain mutations on antibiotic outcome, such as mice incapable of respiratory burst.
Recently, Dr. Conlon collaborated with Duke University to examine Escherichia coli’s in-patient evolution to become more tolerant and its ability to produce persister cells. They found that bacteria can have genotypic changes that do not lead to resistance, but to a high-persister phenotype.4 Dr. Conlon and his team continue to study other mutants that appear clinically in S. aureus and E. coli. Additionally, they are testing for drivers of treatment failure on both the host and bacterial sides. They look at genetic variability in the S. aureus and host through mouse

Figure 2. Antibiotic resistance tests; the bacteria (Escherichia coli) in the culture on the left are sensitive to the antibiotics contained in the white paper discs. The bacteria on the right are resistant to most of the antibiotics. Figure courtesy of Dr Graham Beards via Wikimedia Commons.
infection models, observing how different mouse genetic backgrounds lead to changes in S. aureus variance and antibiotic susceptibility. Other projects look at various components of the innate immune response in relation to S. aureus. The Conlon Lab focuses their work on advancing the field of antibiotics and improving antibiotic efficacy.
Antibiotics are one of the biggest breakthroughs of the 20th century. Resistance and tolerance are the two prominent issues facing it. Dr. Conlon’s work focuses on studying and combating the lesser known, but just as dangerous, antibiotic tolerance. Dr. Conlon’s and his team’s ultimate goal is to strengthen antibiotics and reduce the contribution of tolerance to treatment failure and mortality rates. They eagerly anticipate conducting clinical trials as part of their future plans.1 Dr. Conlon’s research holds promise for significantly impacting the future of healthcare and antibiotic treatment and his team is well on its way to achieving its goal.
1. Interview with Brian P. Conlon, Ph.D. 02/05/2023
2. Fisher, R. A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nature Reviews Microbiology 2017, 15 (8), 453–464. https://doi.org/10.1038/ nrmicro.2017.42.
3. Rowe, S. E.; Wagner, N. J.; Li, L.; Beam, J. E.; Wilkinson, A. D.; Radlinski, L. C.; Zhang, Q.; Miao, E. A.; Conlon, B. P. Reactive Oxygen Species Induce Antibiotic Tolerance during Systemic Staphylococcus Aureus Infection. Nature Microbiology 2019, 5 (2), 282–290. https://doi.org/10.1038/ s41564-019-0627-y.
4. Parsons, J. B.; Sidders, A. E.; Velez, A. Z.; Hanson, B. M.; Angeles-Solano, M.; Ruffin, F.; Rowe, S. E.; Arias, C. A.; Fowler, V. G.; Thaden, J. T.; Conlon, B. P. In-Patient Evolution of a High-Persister Escherichia Coli Strain with Reduced in Vivo Antibiotic Susceptibility. Proceedings of the National Academy of Sciences of the United States of America 2024, 121 (3). https://doi.org/10.1073/pnas.2314514121.

As humans, our molecular machinery is more concerned with preventing our embryos from developing into Duke fans than preventing them from eating their way out of us. However, for the self-fertilizing roundworm Caenorhabditis elegans (C. elegans), a mutation can result in their offspring hatching inside them and consuming their way out, a phenomenon known as a "bag of worms.” To prevent such an unfortunate fate, it’s important to these tiny worms’ survival and fitness to make decisions about where to allocate their energy and resources during reproduction and development. These decisions are highly controlled by regulatory processes and evolutionally conserved genes and pathways, making C. elegans a great model system to study how humans develop and where it can go awry.
“I started out just being very interested in how instructions in the genome turn into phenotypes in organisms,” recalls Dr. Kacy Gordon, the principal investigator of The Gordon Lab.1 During her undergraduate years at Dartmouth, Dr. Gordon became passionate about discovering the origins of life through a developmental and evolutionary perspective. She decided to pursue those passions in graduate school at the University of


Chicago by studying highly conserved DNA regulatory regions among different organisms. Dr. Gordon became an expert at using genetic techniques to research key molecular players involved in development and how they become integrated within the genome throughout evolution. However, it wasn't until her postdoctoral position at Duke that Dr. Gordon found her true love: developmental cell biology in C. elegans
“I wanted to use C. elegans to understand mechanistically what happens at the cellular level when genes are turned on or off,” Dr. Gordon remarks.1 Her curiosity led her to the lab benches of the University of North Carolina at Chapel Hill where she opened her own lab dedicated to studying the function of stem cells during development.
Stem cells are the earliest type of cell within a cell lineage. They exist in an undifferentiated state, meaning they don’t have a specialized function. Stem cells play a central role in both tissue repair and development. In particular, germ stem cells, which give rise to sex cells, are crucial for reproduction as they ensure the transmission of genetic information from one generation to the next. Thus, germ stem cells sit at the crossroads of developmental and evolutionary biology, and understanding its regulation can unlock the mystery of how life came to be.
Dr. Gordon investigates how C. elegans germ stem cells influence their microenvironment, known as the stem cell niche. The germline stem cell niche provides ligands, which are small molecules that bind to receptors to initiate a cellular response. These ligands support stem cell proliferation and renewal while

Figure 2. Brightfield image displaying the Y-shaped gonad bifurcation in ced-12 mutant worms (right). WT on the left which displays normal U-shaped gonad. Image courtesy of Noor Singh.
preventing them from differentiating into sex cells. During C. elegans gonad development, which is the organ that houses the germline, two symmetric U-shaped gonad arms begin to extend out in opposite directions. It first moves away from the front of the midbody, where the vulva is located, then makes a U-turn onto the backside toward the midbody opposite from where it originated (Fig. 1). At the forefront of this pivotal developmental process, termed organogenesis, are the distal tip cells (DTC). The Gordon Lab specifically focuses on these distal tip cells because they serve not only as the stem cell niche, but also as a mass leader of stem cell migration. As the worms reach reproductive adulthood, their germ cells divide and move further away from the influence of the DTC, switching from mitosis to meiosis. Without the DTC, these germ stem cells will begin to ectopically differentiate into oocytes, or unfertilized eggs, and can soon become self-fertilized—a bag of worms best left unopened.
They coined this novel phenotype as “gonad bifurcation.”2 Interestingly, the severed and smaller branch of DTC remain competent to lead cell migration despite not having a nucleus. It was observed that these “zombie cells” continue to lead the migration of stem cells only in the presence of specific ligands provided by the stem cell niche (Fig. 3).2 Rac1 is involved in regulating the actin cytoskeleton by creating protrusions at the leading edge of migrating cells. These protrusions form adhesive complexes with their environment, which facilitate forward movement.3 However, the DTC don’t actually form these protrusive extensions; instead, they use a separate smooth leading-edge mechanism to move. Therefore, when Dr. Gordon and her team assayed for DTC migration in Rac1 mutants, they were shocked to see a bifurcated gonad, a seemingly unrelated phenotype to Rac1 function.
“I wanted to use C. elegans to understand mechanistically what happens at the cellular level when genes are turned on or off.”
C. elegans are translucent, which allows for easy visualization of their DTC with the expression of a fluorescent protein. This is how Noor Singh, a Ph.D. candidate in The Gordon Lab, stumbled upon an unexpected phenotype in the DTC migration pattern when investigating different mutants. The change in DTC migration was caused by a mutation in a gene called ced-10, which is the human equivalent of the Rac1 GTPase gene in C. elegans. Rac1 belongs to a family of proteins known as Rho GTPases, which act as molecular switches that control the organization of the cell's cytoskeleton and play a crucial role in cell migration.2 “We discovered it by accident!” Dr. Gordon says.1
Upon loss of function of ced-10, the DTC continues to lead the migration of the soon-to-be germ cells, but instead of getting to its final destination in one piece, it breaks apart (Fig. 2).

“It’s important to have a plan, but it also pays to be flexible, and if an unexpected and interesting phenotype presents itself, it would be a shame not to follow up on it,” Dr. Gordon explains.1 The next step in this story is to further elucidate Rac1’s seemingly uncoupled but essential role in DTC migration. Furthermore, the development of mammalian lung, kidney, mammary, and salivary glands use a smooth leading-edge mechanism that is similar to DTC.4 However, the developmental processes of these organs remain poorly understood. Therefore, investigating this mysterious phenotype can potentially shed more knowledge into how these organs develop. As Dr. Gordon emphasizes, “You can develop a new hypothesis if you just look carefully enough at what phenotypes manifest upon different gene knockdowns.”1 Often, the answers are right in front of us, waiting to be uncovered.
1. Interview Kacy Gordon, Ph.D. 02/16/2024
2. Noor Singh, Karen Jian Li, Kacy Lynn Gordon. “Getting there in one piece: The Rac pathway prevents cell fragmentation in a nonprotrusively migrating leader cell during organogenesis” BioRxiv. 2023
3. Hanna, S. & El-Sibai, M. “Signaling networks of Rho GTPases in cell motility.” Cell Signal 2013
4. Ewald, A. J., Brenot, A., Duong, M., Chan, B. S. & Werb, Z. “Collective Epithelial Migration and Cell Rearrangements Drive Mammary Branching Morphogenesis.” Dev. Cell. 2008

Evolution is the concept that every living organism developed from other once-living organisms. It sounds simple enough, but the process of evolution is quite complicated. Some species evolve in a few days, while some others take millions of years. Some evolutionary changes are striking, while others can be practically invisible. The only constant? Everything changes.
It is up to scientists to determine how these changes occur. Dr. Maria Servedio is a professor of evolutionary biology in the Biology Department here at the University of North Carolina at Chapel Hill.1 Her research specializes in investigating evolutionary processes through a mathematical modeling lens.
The Servedio Lab focuses on three topics: speciation, sexual selection, and learning. Speciation, according to Dr. Servedio, is the “process of one species splitting into two”. As one species evolves, there are subgroups of organisms within that species that develop their own unique characteristics and evolve into a separate species. According to Dr. Servedio, “everything on Earth ultimately resulted from a speciation process.” In its most basic form, speciation usually occurs when there is a geographic barrier. A common example is Darwin’s Galapagos finches. Scattered across the Galapagos islands are different species of finch, all evolving from one ancestor finch. They have various beak sizes depending on the food source available – shorter beaks to eat tiny seeds, and larger beaks to eat insects. Although this pattern applies to geographic speciation, this could also apply to speciation on a single island. Dr. Servedio focuses on
speciation in this context: working on models that predict how and if speciation can even occur in certain environments. For example, Figure 2 predicts if female birds prefer certain traits in males which developed based on different environmental pressures. This graph comes from Dr. Servedio’s recently developed hypothesis called the “ecological stage hypothesis” which explores speciation in a geographic context.2
 Dr. Maria Servedio
Dr. Maria Servedio
Modeling speciation involves sexual selection, where individuals choose their mates based on certain traits or behaviors that are attractive to the opposite sex, like feather coloration, songs, and calls. To symbolize a female making a mate choice, Dr. Servedio uses a peacock, as physical traits in male peacocks, like tail color and length are attractive to female peacocks.3 When sexual selection varies in a geographic context, it complicates evolution. “If an organism can change who they are likely to mate with just based off their encounters, then things like who is around in the population can flip the mating preferences drastically,” explains Dr. Servedio.1 There is another layer to the process: mating preferences can be learned. Dr. Servedio noted that it is “hard to imagine” a world where “mating preference have no cultural influence.” Even humans have learned preferences; think of beauty trends, or standards that influence the way we choose our partners.1 Learned attraction applies to animals and introduces numerous evolutionary possibilities. For example, if an individual had a firm rule that they would only mate with individuals that resembled their parent, then that would change who the individual ultimately mated with. Or if they only mate with individuals they prefer, but

Figure 2. Model explaining how reproductive isolation evolves based on female trait preference. Image courtesy of Dr. Maria Servedio.
learned these preferences from their parents, this can create a lot of differentiation. Especially if the individual’s parents were extremely unusual. Ultimately, “learned” mating can create more differentiation within a species. In an evolutionary sense, Dr. Servedio explained that learning can create “large and slightly unpredictable affects,” which makes it the perfect process to mathematicise.1 Dr. Servedio wanted to better understand learning in a speciation context. Many questions influence her research: what if an organism’s preferences are learned, instead of genetic? Does reproduction get harder or easier? How does learning change the way speciation works? Dr. Servedio’s use of models in her research is abstract. Instead of using data from any one species, she uses math to test “verbal chains of logic,” which are ideas that scientists come up with when they are generating hypotheses.1 From these hypotheses, Dr. Servedio builds population genetic models that test how logically sound these various predictions are. Currently, Dr. Servedio is working towards a better understanding of evolutionary learning and has developed a novel hypothesis for how sexual selection might work. Her “inferred attractiveness” model explains how some species observe the choices of others before they make a choice. Observing these choices is complicated, however, as “[individuals] are not omniscient observers that know exactly what another individual is looking at [when they choose a mate].” An observer female might think that another female is attracted to her mate’s coloration, but the female could be attracted to her mate’s tail length, instead. The observer female, however, would only be influenced by her perception of what the female is seeing in her mate. This “error in observation” can create complex patterns in male trait frequencies that can be explored through mathematical situations. Visualizing the various possibilities through a mathematical lens is crucial to understanding all the pathways evolution can take.1 Dr. Servedio’s model, which depicts species trait frequencies, can be used to visualize preferences and interpret changes based off graphs, especially when making research conclusions.4 Math based “proof of concept” models that test predictions can make “powerful insights that scientists are not going to
get any other way.”1
When it comes to evolution, there is a lot more to learn! There is a misconception that all science needs to have a direct benefit—such as bettering healthcare or solving climate change. Evolution is considered “basic science”— meaning science conducted solely for knowledge’s sake. For Dr. Servedio, “there is nothing wrong with questions that are only of intellectual interest”, as applications of knowledge often emerge in ways scientists did not initially consider.1 This evolutionary research is important because it has conservation implications. As species evolve due to the changing climate, it is essential for scientists to understand why and how these populations are changing. According to Dr. Servedio, “science in general is about learning as much as we can,” and evolutionary research does just that: it provides important insight into changes within the world around us.

Figure 3. Male trait and female preference frequencies model. Image courtesy of Dr. Maria Servedio.
1. Interview with Maria Servedio, Ph.D. 01/22/24.
2. Boughman, Janette W., Servedio, Maria R., The ecological stage maintains preference differentiation and promotes speciation, Ecology Letters, 2022, 25(4). DOI: 10.1111.
3. Servedio, Maria R., Burger, Reinhard, The counterintuitive role of sexual selection in species maintenance and speciation, PNAS, 2014. DOI: 10.1073.
4. DuVal, Emily H., Fitzpatrick Courtney L., Hobson Elizabeth A., Servedio Maria R., Inferred Attractiveness: A generalized mechanism for sexual selection that can maintain variation in traits and preferences over time, PLOS Biology, 2023, 21(10). DOI:10.1371.
The tapestry of life is an elaborate web of cellular wonders meticulously woven throughout the human body. This symphony of existence relies on the seamless function of every cell and tissue. Within this complex arrangement, epithelial tissue stands out for its critical role. At the forefront of unraveling these mysteries is Dr. Peifer and his crew, a team of curious minds dedicated to understanding how our cells develop and interact with one another. Their goal is to uncover what drives cell shape change and cell motility, especially those involving myosin arrays and cytoskeletal assembly.

Dr. Peifer's enthusiasm is infectious when he discusses his lab's motivations.
"Our main interest right now is understanding how cells change shape or move, how you build tissues and organs, and how you do that with tissues and organs tearing themselves apart," he says.1 The team delves into the complexities of cellular adhesion and function, exploring the nuances of signaling, adhesion, and motility that are essential for maintaining healthy tissues.
Dr. Peifer, reflecting on the complexities of his research, shares, "So for cells to change shape and move, you need to generate force. And then the cytoskeleton is usually involved with that, and we focus on the actin and myosin cytoskeleton".1 This statement captures the essence of their findings, highlighting the critical role of myosin arrays in driving the morphological changes necessary for tissue development. Dr. Peifer discusses the intricate balance between cellular adhesion and mobility, "You need to do that in ways that are dynamic. And you need to be able to pull hard on those junctions and do that without ripping the cells apart".1 This delicate interplay is at the heart of their research, underscoring the innovative approaches the team employs to unravel the mysteries of cellular behavior.
Among the myriad of projects undertaken by Dr. Peifer's lab, one
particularly stands out for its in-depth exploration into the cellular mechanics essential for tissue and organ formation. This project, elaborated in their recent publication, examines the crucial role of micron-scale supramolecular myosin
“Our main interest right now is understanding how cells chance shape or move, how you build tissues and organs, and how you do that with tissues and organs tearing themselves apart"
arrays in the remodeling of epithelial tissues. The research provides evidence that these myosin arrays, contrary to previous beliefs, play a key role in assembly of the cytoskeleton at cell-cell adherens junctions (AJs), rather than this being driven only through actin nucleation and polymerization mediated by machines like Arp2/3, formins, or Ena/VASP proteins [Figures 1 and 2]. Surprisingly, "inhibiting Arp2/3 or formins reduced actin levels at cell junctions," yet this did not prevent the formation of myosin arrays and tightly bundled actin, suggesting a novel understanding of actin organization at mature adherens junctions [Figure 1].2 "Now, we're really thinking again about how cells change shape or move, and how you build tissues and organs," Dr. Peifer
shares, emphasizing their commitment to unveiling the complexities of cellular dynamics.1 The study not only enriches our understanding of cellular dynamics but also illustrates the lab's broader interest in how tissues and organs are sculpted during embryogenesis.
This pivotal discovery indicates that the assembly of these myosin arrays, which are essential for the concerted movement and structural integrity of cells within tissues, might be driven by mechanisms previously underappreciated. The study's findings challenge the traditional emphasis on the singular importance of actin polymerization machines, revealing that "individual activities of Arp2/3, formins, or Ena/VASP proteins are not essential for assembling supramolecular actomyosin structures at the ZA".2 This insight opens new avenues for understanding how cellular forces are regulated and orchestrated during tissue development, highlighting a potential collective action of multiple actin polymerization machines and the importance of further investigating the roles of actin-binding proteins in concert with myosin to organize and bundle actin filaments.2
The lab's success is partly due to their access to structured illumination microscopy and CRISPR, allowing them
to manipulate gene and protein structure and then to observe and decipher cellular behaviors with unmatched clarity. "We're fortunate on this campus to have always been at the cutting edge of microscopy," Dr. Peifer acknowledges. This highcaliber equipment is crucial for their groundbreaking discoveries in capturing embryo development and migrating muscle precursors.
However, all of these techniques are secondary to the Drosophila, an organism with unparalleled advantages. Dr. Peifer points out, "The biggest advantage of the fruit fly is that we know a lot. Just the fact that people have been working on it for 120 years means we're at the cutting edge." This extensive background supports their hypothesis testing and the observation of cellular activities in vivo.
Yet, the path of research is fraught with challenges. Dr. Peifer openly discusses the hurdles of moving beyond traditional model organisms and deciphering protein interactions at the microscopic level. "We're trying to fill that gap, coming from the bottom using new forms of electron microscopy and coming down from the top, trying to get even better resolution," he reveals.
Despite the challenges, the thrill of discovery propels the team forward. Their work is enhancing our fundamental
understanding of life at the cellular level and holds promise for future medical breakthroughs that relate to cell-cell junctions and cytoskeletal regulation. Through meticulous research and a spirit of inquiry, Dr. Peifer's lab leads the way in uncovering the mysteries of cellular and tissue biology, paving new paths in science and medicine.
1. Interview with Mark Peifer, Ph.D. 2/23/2024
2. Yu-Kemp, H.-C.; Szymanski, R. A.; Cortes, D. B.; etc. Journal of Cell Biology 2021, 221 (1): e202103074

Space exploration captivates the minds of many, drawing us into a realm of limitless possibilities for humanity’s future. With its allure of inhabiting distant worlds and uncovering the mysteries of the cosmos, space travel captivates our imagination like no other scientific endeavor. Yet, this journey is not without its perils. The harsh realities of radiation exposure and weightlessness pose significant challenges to the human body. However, as our understanding of cellular aging advances, hope emerges on the horizon. Pioneering research from the Schisler Lab offers promising solutions to counter the effects of space travel on astronauts, paving the way for safer voyages beyond our planet. Intriguingly, insights gained from astronauts’ experiences also hold the potential to benefit patients here on Earth, bridging the gap between outer space and medical advancements on our home planet.
As space travel expands, it has become a priority to ensure that long duration space travel be completely sustainable to the human body in order to realize any of humanity’s space ambitions. Outside of Earth’s safe bubble of atmosphere however, astronauts face unique experiences that stretches the boundaries of human cell resilience. The physiological changes experienced during spaceflight, including weightlessness and exposure to ionizing radiation, lead to heavy mechanical stress on cell bodies, DNA degradation, and increased stress on the mitochondria. These effects can be devastating to the human body and
By Morcos Saeeddepending on the types of cells affected, they can lead to neurodegenerative disease like Alzheimer’s disease, cardiovascular failure, or even cancer. NASA’s efforts in long-term spaceflight focus on developing what they call “countermeasures” to mitigate any and all of the issues that stand in the way of long- term safe spaceflight. Key among these countermeasures is understanding the effects of microgravity and cosmic radiation on astronaut health. By studying the molecular and cellular responses to these stressors, researchers aim to develop countermeasures to reduce or even completely end potential health risks for astronauts. As such, understanding the impact of space travel on mitochondrial and cellular health is particularly crucial, as it underscores the importance of maintaining cellular resilience in extreme environments like space.
Far from space and back here on earth, humans and other organisms are born and grow. During this process, the cells composing their body continuously duplicate on an almost constant basis providing support and growth for their bodies. Some of these tend to replicate only during the early stages in life and then cease to replicate. These cells include neurons and cardiomyocytes which specialize in vital energy-hungry organs such as the brain and heart, respectively. As we get older, however, the reliability of these cells begins to dwindle, similar to car aging; there is a lot of wear and tear involved in cellular aging. Our body
possesses mechanisms and proteins specialized in protein folding necessary for complex proteins as well as destroying dying or potentially dangerous structures. Among these, a protein called CHIP is especially interesting for researchers because it can perform both the functions of protein folding and marking cells for degradation. The stress caused by cellular aging is associated with multiple age-related diseases such as Alzheimer’s disease, cardiovascular disease, and even cancer. Thus, it is useful to find models

Dr. Schisler, a leading expert in the field of cellular aging and an Dr. Jonathan Schisler
and data from situations where cells have been exposed to extreme physiological conditions leading to cellular stress and mitochondrial failure. This will improve the study of how to use proteins like CHIP to combat the effects of cellular aging and find a therapeutic approach to tackle many issues related to cellular aging’s disease implications. Such conditions can be induced on vitro cells in a lab unless they can be found somewhere else, for example in space.

Figure 1. An attempt at remodeling the fibers of cardiac tissue after exposure to galactic cosmic radiation and solar irradiation. This is the micrsocope figure obtained from the analysis on mice tissue. No changes were observed. Figure courtesy of Beheshti et al.
Associate professor in the Department of Pharmacology at the University of North Carolina at Chapel Hill School of Medicine, emphasizes the significance of utilizing models to gain insights that can inform human health. His research focuses on understanding the molecular mechanisms underlying cellular stress responses, particularly in the context of aging and disease in non- regenerating cells like neurons or cardiomyocytes.
Drawing parallels between the resilience of neurons and cardiomyocytes, Dr. Schisler highlights the critical role of DNA integrity and replication fidelity in maintaining cellular function. His work on the protein CHIP explores its potential as a therapeutic target for aging-related disorders through its unique properties. On the other hand, NASA programs like the STAR program focus can help biomedical researchers who in turn can provide valuable insights and solutions for a safer long term space travel.
In the presence of this win-win situation,
the Schisler Lab has and continues to provide and receive valuable data from the STAR program. The exchange highlighted a correlation between cellular aging effects and mitochondrial stress expressions as research continues on how to benefit from the provided information to develop a therapeutic approach for earth age related diseases. Schisler’s perspective underscores the interdisciplinary nature of cellular resilience research, where insights from mouse models, space research, and clinical observations converge to advance our understanding of human health and resilience. As he aptly puts it, “I like using models and I like being able to get information from models that could inform us on how the mechanism works… ultimately we are here for human health and sort of the non-astronauts out there and we are interested in treating in particular monogenic disorder… It is another model system where we are getting more information on neuro resilience response to stress that may turn out to be useful for other diseases on planet earth.”
In conclusion, the integration of pharmacology and space research, along

Figure 3. Guide RNA to target CHIP S20/S24 region, and the replacement RNA that includes a silent mutation of the PAM sequence to further enhance homology directed repair. It bascially showed how a mutation can induce myocardial infraction and it is done clinically in the lab similar to pharmachological processes done regularly at the Schisler lab. Figure courtesy of Ranek et al.

Figure 2. NASA STAR program logo. Image courtesy of NASA.
with the perspectives of experts like Dr. Schisler, provides a comprehensive approach to unraveling the complexities of cellular resilience and mitochondrial health. By elucidating the molecular mechanisms underlying cellular stress responses, researchers aim to develop novel therapeutics and interventions to promote human health and resilience both on Earth and beyond.
1. Interview with Jonathan C. Schisler PhD, 10/12/2023
2. Ranek, M. J., Oeing, C. U., SanchezHodge, R., Kokkonen-Simon, K. M., Dillard, D., Aslam, M., Rainer, P. P., Mishra, S., Dunkerly-Eyring, B., Holewinski, R. J., Virus, C., Zhang, H., Mannion, M. M., Agrawal, V., Hah n, V. S., Lee, D. I., Sasaki, M., Van Eyk, J. E., Willis, M. S., . . . Kass, D. A. (2020). CHIP phosphorylation by protein kinase G enhances protein quality control and attenuates cardiac ischemic injury. Nature Communications, 2020, 11(1). https:// doi.org/10.1038/s41467-020-18980-x
3. Beheshti, A., Paar, V., Jiang, S., Enriquez, A., Kim, J., Brunetta, H. S., Muratani, M., Kubik, A., Allen, N., Blaber, E. A., Overbey, E., Altınok, S., Sanchez-Hodge, R., Oswalt, L., Kaufman, B. A., Mori, M. A., Motloch, L. J., Mason, C. E., Schisler, J. C., & Jirak, P. (2023). Countermeasures for cardiac fibrosis in space travel: It takes more than a towel for a hitchhiker’s guide to the galaxy. Research Square, 2023. https:// doi.org/10.21203/rs.3.rs-2351744/v1

As the climate continues to change, it is expected that there will be a difference in food production due to rapidly changing environments.1 To maintain a stable crop yield, we must begin to examine how to best enhance crop survival. A closer look at plant immune responses may be the key to this goal.
Dr. Jeff Dangl is a professor of biology at The University of North Carolina at Chapel Hill holding the John Couch distinguished professorship and a Howard Hughes Medical Institute investigator.2 Originally a modern literature major at Stanford University, Dr. Dangl discovered his interest in biology when he worked in a mouse immunology lab as an undergraduate. His interest in the niche of plant immunity began after reading a paper on plant cell recognition and gene transcription while working toward his Ph.D. Along with the paper, he recognized that plant breeders have long worked to develop pathogen-resistant plants using genetics, but that in the mid1980s, there had been no tools to discover the genes that rendered plants resistant to pathogens. To him, this indicated a link between the genome and disease
resistance, and with these two themes in mind, he began to consider the possibility of pathogen resistance as a set of traits directed by a receptor system within the plant cells.
Much like humans, plants have an immune system that helps protect them from harmful pathogens.2 Instead of circulating immune cells, a twostep system helps protect plants from bacterial invasion. Primarily, the cell surface receptors recognize non-self bacterial or fungal components which trigger an immune response inside the cell. This response alters specific gene expression to code for proteins that inhibit further growth of the pathogen. However, the pathogen can adapt its own

proteins to silence this immune response and colonize the plant without triggering inhibition. As a second immune response, receptors inside the cell recognize these pathogen proteins and re-initiate the correct immune response. As a result, plants and microbes are constantly adapting and evolving to counteract each other’s invasion and response techniques.
For effective microbe infiltration, there are four major steps that it must undergo: colonizing, persisting, thriving in a changing environment, and multiplying.2,3 In short, the microbe must be able to enter a plant organ such as a leaf or a root, survive the immune response, continue to survive in a natural environment alongside the plant, and carry out its designated functions. As a result of the two-step immune system, it can be incredibly difficult for microbes to achieve even the first two goals.
The Dangl Lab focuses on several projects related to this central theme of plant-microbe interactions by combining the ideas of immune receptors and beneficial microbe relationships within the roots.2 The lab studies Arabidopsis thaliana (commonly known as thale cress, a close relative of mustard and Dr. Jeff Dangl
cabbage), as a model species. A. thaliana contains a low number of chromosomes and a small, compact genome, but a gene number roughly equal to common crops such as rice and corn. Due to this similarity, outcomes from experiments involving the species can easily be applied to other crops. Additionally, it has 5 to 6-week generations, and can be infected by pathogens related to those that target its crop relatives. As a result, A. thaliana has become a primary model for experimentation in pathology, genetics, and disease resistance.
One of the lab’s primary goals is to observe the effects of different microbes on plants in varying environments.2 Within their studies, the lab observes the changes in the growth of plants in different simulated environments. Ideally, a beneficial microbe would help a plant survive within changing environmental conditions. For example, a microbe may be able to help a plant survive through a drought season, block infection, or provide it with an essential nutrient that it lacks.2,3 An example of this type of research includes studying the impact of various bacterial strains introduced to plants in three different environments as seen in Figure 2.4 This figure depicts the lab’s research into the change in A. thaliana biomass as a result of different combinations of microbes and environmental stressors.
Another goal of the Dangl Lab is to understand the process of plant immune

response to microbes and identify the genetic determinants of plant disease resistance.2 There is great diversity in the genes involved in the immune response of A. thaliana; roughly 600 genes code for external receptors and 500 for internal recognition proteins. Furthermore, triggering these immune responses alters the transcription in around 2,000 genes. Because of the relatively large number of genes involved in immunity, Dangl’s work emphasizes studying resistance at the DNA level.
As Dangl puts it, most biological research and laboratory testing related to plant biology have long-term goals in human interaction and environmental health.2 He states that the long-term goal of research in the field of plant immune systems and microbiomes lies within agriculture. He believes that by identifying how plant immune systems function, researchers can harness this understanding to enhance food production in the future.
One of the primary veins of focus he expects to see in the future is an alternative to pesticide use.2 Due to the use of homogeneous farming methods— the farming of a single strain of a single species per plot of land—pathogens that wipe out entire crop yields are not uncommon. Climate change introduces new pathogens into areas where they could not previously thrive, adding to the possibility of crop failure. Dangl claims that research into plant immunity can help shed light on how to avoid these potential dangers through predictive breeding and genetic engineering.
Similarly, he believes that the use of specifically developed microbes may be the next step in aiding crop growth and survival in a changing climate.2 Alongside predictive breeding, pathogen-inhibiting microbes can be used as an alternative to pesticides. Microbes are already ‘natural’ to the plant’s system, and through DNA manipulation, researchers can develop single microbes that combat disease, the effects of drought, and limited nutrients.2,3 While this goal is more distant, Dangl believes that microbial support can become a primary resource to improve crop stability. Just as plants and bacteria perform their complex back-and-forth, the world must adapt its practices to ensure a lasting relationship between humans and a perpetually changing global environment.
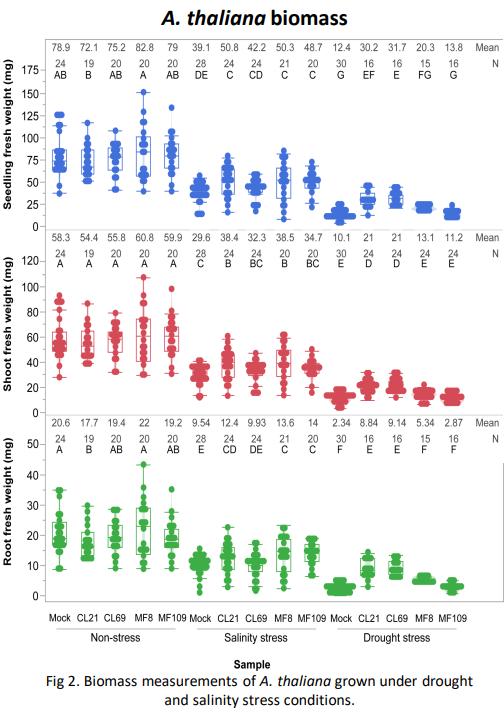
Figure 2. Biomass measurements of A. thaliana grown under drought and salinity stress conditions. Figure courtesy of Audrey Thomas.
1. Gowda, P.; Steiner J.L.; Olson C.; Boggess M.; Farrigan T; Grusak M.A. Agriculture and Rural Communities. In Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment, Volume II. Reidmiller, D.R., Avery C.W., Easterling D.R., Kunkel K.E., Lewis K.L.M., Maycock T.K., Stewart B.C., Eds.; U.S. Global Change Research Program: Washington, DC, pp. 391–437. doi: 10.7930/NCA4.2018.CH10
2. Interview with Jeff Dangl, Ph.D. 2/08/2024.
3. Russ, D.; Fitzpatrick, C.R.; Teixeira, P.J.P.L.; Dangl J.L. Deep discovery informs difficult deployment in plant microbiome science. Cell. 2023, 186, 4496-4513. https://doi.org/10.1016/j. cell.2023.08.035
4. Thomas, A.; Ibeanu, T.; Patel, K.; Eida, A.A.; Dangl, J.L. Individual syncom-34 bacteria strains demonstrate plant rescue under drought and salinity stress in Arabidopsis thaliana. Naval Academy Science and Engineering Conference, United States Naval Academy 2023
 By Preston Szczesniak
By Preston Szczesniak
Is wildlife research only focused on animals? No, it also includes plants! This is exactly the type of research that Dr. Damon E. Waitt leads at the North Carolina Botanical Gardens near the University of North Carolina at Chapel Hill campus. Dr. Damon E. Waitt is a Professor of the Practice in the Department of Biology at the University of North Carolina at Chapel Hill. He is also the Director of the North Carolina Botanical Gardens. Dr. Waitt received his Bachelor of Science in Biology from Tulane University, his Master’s degree in Botany from Louisiana State University in Baton Rouge, and his Ph.D. in Botany from the University of Texas at Austin.1

He has held many roles in leading initiatives for plant conservation and research, including the following: acting as a Senior Director and Botanist at the Lady Bird Johnson Wildflower Center from 2001-2015, contributing to the Invasive Species Advisory Committee of the National Invasive Species Council, assuming the role of President of the Texas Academy of Science, leading as the founding president of the Texas Invasive Plant and Pest Council, and serving as both the Chair of the North Carolina Plant Conservation Program and the interim President and CEO of the Center for Plant Conservation.

Figure 1. One of the research projects Dr. Waitt heads is the UNC-Chapel Hill Herbarium’s research, which recently produced a plant research and conservation publication titled Flora of the Southeastern United States, covering the flora of the states shown in this map. Figure from North Carolina Botanical Garden Website.
As the Director of the North Carolina Botanical Gardens, Dr. Waitt leads research initiatives in plant conservation and the work conducted by the garden’s staff members. For instance, he facilitates research conducted by the UNC-Chapel Hill Herbarium (plant collections), which he says is leading research in “…fundamental plant taxonomy and systematics”.2 In fact, according to Dr. Waitt, around a few months ago, this



Figure 2. Dr. Waitt oversees the Plant Conservation Program’s research that it conducts in natural areas, which are shown in green on this map, leading to successful projects in plant research and conservation under the Plant Conservation Program at the North Carolina Botanical Gardens. Figure courtesy of Dr. Waitt.
work described ten new plant species! One of the publications produced by the UNC-Chapel Hill Herbarium in 2023 was the Flora of the Southeastern United States, which covers the plant biodiversity of regions in the southeastern United States, specifically in the states shown in Figure 1.3 These research programs facilitated by Dr. Waitt have become established primary sources for individuals seeking information about the plant biodiversity of the southeastern United States. The UNC Herbarium’s research also ultimately led to the development of the FloraQuest app. Specifically, it enables users to look up information for any of the 5,800 documented plant species found within the regions detailed in Figure 1, teaching its users facts about the plant species, such as its geographic distribution in the regions illustrated in Figure 1. You don’t even need an internet connection to use it!
Additionally, Dr. Waitt oversees the Plant Conservation Program at the North Carolina Botanical Gardens, which is focused on ex situ conservation, the conservation of a species outside of its native habitat, and in situ conservation, the conservation of a species inside its native habitat. This includes rare plant species recovery, reintroductions, seed banking, and studying their biology and ecology. One aspect of the Plant Conservation Program that Dr. Waitt facilitates initiatives in is plant research and conservation in various spaces managed by the North Carolina Botanical Gardens, such as Penny’s Bend, the Piedmont Nature Trails, and the Mason Farm Biological Reserve. In fact, the Mason Farm Biological Reserve is the site for longterm research projects such as a global carbon storage study, as well as a long-term bird breeding survey. These areas, along with other spaces that the North Carolina Botanical Gardens oversees, are shown in the map present in Figure 2.
Another element of the Plant Conservation Program that Dr. Waitt leads is the Plant Materials of the Atlantic Southeast
program, which involves collecting seeds from native plant species to preserve plant biodiversity by ensuring the restoration of these species in their native habitats. As of 2019, the program has collected seeds from 245 native plant species across 626 total collections. Dr. Waitt also facilitates the Plant Conservation Program’s rare plant recovery programs, reintroducing native plant species into habitats that they formerly inhabited. An example of this is their efforts to reintroduce 4 specific plant species named Lilium pyrophilum (Figure 3), Astragalus michauxii, Amorpha georgiana, and Lysimachia asperulifolia
Finally, Dr. Waitt leads the Plant Conservation Program’s efforts in preserving the genomes, or the entire genetic material, of native plant species in what is known as seed banking. This is an ex situ conservation effort that helps promote plant research and conservation by storing the genetic material of these native plant species. Under Dr. Waitt’s leadership, the Plant Conservation Program works with other organizations such as the Center for Plant Conservation to seed bank native populations of Mountain purple pitcher plants, Piedmont fameflowers, and Blue Ridge bog huckleberries (Figure 4).
Ultimately, the goal of the research that Dr. Waitt manages at the North Carolina Botanical Gardens is to study and protect native plant species. The research he facilitates leads to specific efforts in successfully conserving the natural world. Furthermore, he states that the research he directs is critical in addressing the biodiversity crisis not only for plants, but also for other species such as native animal species: “Birds, insects—all sorts of things are going extinct under our very noses.” He hopes that the applied plant research he facilitates at the North Carolina Botanical Gardens will ensure the future of these native plant species so that they can continue to be cherished in their native land.

Figure 4. Mountain purple pitcher plants. Figure from Center for Plant Conservation Website.
1. Dr. Damon E. Waitt, Ph.D.: BIO. Document courtesy of Dr. Damon Waitt.
2. Interview with Dr. Damon E. Waitt, Ph.D. 01/24/2024.
3. North Carolina Botanical Gardens. 2024. Research. Chapel Hill, North Carolina, United States. https://ncbg.unc.edu/ research/.

Each time we step out of our home—whether it be a house, apartment, or dorm—we inhale the freshness of the surrounding air, an automatic act that often goes unnoticed. This routine inhalation and exhalation actually exposes us to a world of pollutants and particulate matter, posing a growing threat to our well-being. The alarming rise in pollution levels over the last few decades has been linked to numerous health concerns, with ozone and particulate matter (PM) emerging as silent culprits responsible for millions of premature deaths globally.1

At the forefront of combating these environmental challenges is Dr. William Vizuete, a distinguished professor in the Department of Environmental Sciences and Engineering at the Gillings School of Public Health at University of North Carolina at Chapel Hill. His impactful work goes well beyond global boundaries to address environmental issues in many places, including Texas, Colorado, Abu Dhabi, Dubai, Brazil, Ecuador, and East Africa. Dr. Vizuete’s dedication to tackling these critical problems has not only fostered strong relationships with policymakers but also earned him international acclaim.
Throughout his undergraduate career, Dr. Vizuete was interested in solving problems using science, inspiring him to pursue chemical engineering. It was during this time that he became intrigued with the long-standing problem of air quality and aerosols. Aerosols are the deadliest air pollutants, and exposure to them can lead to a variety of health issues. Depending on their composition, size, and location, aerosols can either heat or cool the earth’s atmosphere. As Dr.Vizuete
notes, “One of the largest uncertainties in our climate model is the behavior of these aerosols.”1 The unpredictable behavior of aerosols creates one of the largest uncertainties in Dr. Vizuete’s study, making it challenging to predict the effects of these pollutants.
To understand the impact of pollutant exposure on human health, the origins of pollutants, and the entities responsible for their production, air quality models serve as a pivotal tool. According to Dr.Vizuete, “One very important tool for understanding the exposure, sources, predicting the future and how future policy would change air quality, or to understand what communities are more exposed then others, is air quality models.”1 These computational models, based on sophisticated algorithms and scientific data, function as frameworks that facilitate the explanation of complex relationships between atmospheric components, pollutant sources, and their subsequent effects on public health. As such, Dr. Vizuete engineers and applies these models to better demonstrate these relationships and help others understand them as well.
Dr. Vizuete’s research group is split into two different groups. The first group concentrates on improving the air
quality simulations by integrating the latest lab discoveries and findings to create a better model. Creating a perfect model is nearly impossible, but through continuous improvements, this “model development” group leads to a highly effective model. The second group is focused on the application of the air quality model. This involves taking the findings generated by the first group, understanding what the simulations are saying, and advising policymakers, environmental justice advocates, and other scientists on the best course of action.
In one such study relating air quality to health, individual and co-occurring chemicals in the atmosphere were tested to see if they caused modified expression of genes involved in inflammation and cancer-related processes in lung cells. The study focused on pollution in the Houston Ship Channel Area in Texas, which has the biggest number of petrochemical facilities within the United States. Since it is difficult to study single pollutants, as the atmosphere has various chemicals interacting with PM and climate conditions, the goal was to study cell responses to complex gas mixtures similar to those of true atmospheric conditions. Human lung cells were exposed to air pollutant mixtures similar to atmosphere mixtures collected across five days at the University of Houston field site. The results showed differential gene expression in 88 unique genes that were related to 16 unique air pollutants. The biggest pollutant that was identified corresponding with the largest transcriptional response was benzene. Benzene is a recognized Hazardous Air Pollutant associated with the development of cancers and cardiovascular disease. Among the genes that were studied TNF (tumor necrosis factor), p53, and IRF1 (interferon regulatory factor 1)—all known for their roles in regulating cancer processes—demonstrated changes in their expression levels and activity.2
Figure 2. Genes significantly differentiated through benzene exposure. Photo courtesy of Vizuete et al.
immediate free recall and new learning. This neurotoxicity, caused by the gradual shrinking of the brain’s grey matter (a type of tissue in the brain and spinal cord), is linked to elevated Alzheimer’s disease risk, independent of damage to blood vessels in the brain.3
Through all of these findings, we can see the enormous impacts of air pollution on human health. Dr. Vizuete works at all scales of the model—globally, regionally, and locally—to allow for policy improvements in all regions. Through the ECUIPP lab, undergraduate students can perform community-based research on air quality issues in North Carolina highlighting one of many opportunities for environmental research. As the world continues to develop at a rapid rate, there is much more to learn about air pollution and its effects on human health. Dr.Vizuete is one of the many dedicated scientists helping us to do so.
In another study, the effects of exposure to particulate matter (PM) were studied in correlation with Alzheimer’s disease and dementia. The research studied whether PM influences changes in brain structure and accelerates neuropsychological processes, specifically the decline of episodic memory, a marker of Alzheimer’s disease. The study, conducted with older female participants, revealed that PM is associated with greater declines in immediate recall and new learning. Furthermore, the research explored the potential mediating role of increased risk for Alzheimer’s disease linked to PM exposure, demonstrating that long-term exposure is associated with increased Alzheimer’s disease pattern. These findings emphasize the influence of neurotoxicity related to PM2.5 (PM with a diameter less than 2.5 micrometers) which contributes to the early decline of
1. Interview with William Vizuete, Ph.D. 2/9/2024
2. Lauren A. Eaves, Hang T. Nguyen, Julia E. Rager, Kenneth G. Sexton, Thomas Howard, Lisa Smeester, Anastasia N. Freedman, Kjersti M. Aagaard, Cynthia Shope, Barry Lefer, James H. Flynn, Mathew H. Erickson, Rebecca C. Fry, and William Vizuete. Environmental Science & Technology 2020 54 (21), 13807-13816
3. Younan, Diana et al. “Particulate matter and episodic memory decline mediated by early neuroanatomic biomarkers of Alzheimer’s disease.” Brain : a journal of neurology vol. 143,1 (2020): 289-302. doi:10.1093/brain/awz348

Amphibians, including many species of salamander, breathe predominantly across their skin and very little with their lungs – some salamanders have even evolved to be completely lungless. Their skin is very thin and highly vascularized, allowing for this unique mode of skin breathing. Dr. Eric Riddell of the University of North Carolina at Chapel Hill Biology Department compares salamanders to “walking lungs” because of this ability, as they even contain similar receptors on their skin that other animals have only have on the surface of their lungs.1
A recent publication, spearheaded by Dr. Riddell and his graduate student, Issi Burger, focused on understanding the specific trade-off between water loss and breathing in amphibians. Salamanders, like all other terrestrial species on Earth, experience many challenges associated with living on land. One of the most challenging aspects of living on land is staying hydrated, but in order to breathe oxygen, plants and animals must lose water. Humans experience a similar challenge as we exhale much more humid air than we breathe in. This process becomes a complicated relationship of give and take to produce the best evolutionary combination of high oxygen uptake and minimal water loss.1 As organisms hybridize, or produce new species through mating over time,
this relationship may influence the survival of hybrid species in different environments. The researchers in this study looked at how the physiology of the endangered California tiger salamander, a charismatic and imperiled salamander of California, compared to the hybrid tiger salamanders, which are rapidly expanding across California. Their study suggests that hybrid tiger salamanders are so successful because of their unique ability to breathe with relatively little water loss – a trait that the native species does not share. This research could have
 Dr. Eric Riddell
Dr. Eric Riddell
massive implications for the future of amphibian populations through climate change and species adaptation over diverse environments.
Dr. Eric Riddell, the principal investigator on this project, has “always been passionate about the deeper connections within science, especially those that you wouldn’t normally imagine."1 He spent time working on the Galapagos Islands before graduate school, researching the ecological connections between climate shifts and changes in the diet of a seabird (Nazca booby) that caused larger population declines on the islands. It thus makes sense for Riddell to be at the forefront of groundbreaking research on novel ecological connections in physiology. This is what makes Riddell’s new study unique: the use of a new metric of overall respiration performance to study an organism’s fitness with new precision. This new metric is a ratio that compares gas exchange to evaporative water loss –defining an organism's ability to perform energy-intensive activities (as through increased gas exchange) while reducing the amount of water loss necessary for respiration. The Water-Gas Exchange Ratio (WGER) helps standardize these physiological rates. It expresses the rates as a single trait, rather than two separate traits to compare this joined aspect of physiology across species or in this case,
across hybrids of the salamander species.1 The information provided by this metric will help researchers understand why certain hybrid species of salamanders have increased in population so rapidly and can potentially evolve more efficiently than other species over time. This test is a novel way of researching how animals perform in the wild with coupled physiological traits.
The results of the experiment conclude that hybrids exhibited a higher WGER compared to the two parental species, suggesting greater respiration efficiency in hybrids (illustrated in Figure 1).2 The hybrid species breathe more efficiently and are therefore able to adapt and expand their geographic range at a high speed. Additionally, individuals found to have higher resistance to water loss had lower metabolic rates, supporting the potential trade-off discussed between these two particular traits in hybrids. California tiger salamanders, however, showed no trade-off between metabolic rate and water loss using this research metric. This indicates that these two properties may be dissociated in the species, suggesting why their fitness falls behind hybrids. The overall increased WGER in hybrids gives a plausible explanation for rapid hybrid growth over the California terrain, as well as more evidence of hybrid vigor (these hybrids outcompeting their parent species) in a broad system.
Overall, these results provide greater insight into the mechanisms behind hybrid fitness and highlight the importance of multi-trait analyses in studying organism physiology. Riddell’s study potentially provides a mechanistic explanation for the expansion of hybrids

future of biodiversity and competition between species. Riddell looks to expand on this research in the near future by examining the evolution of these traits: how they coupled together to create such interesting and vital trade-offs, and how we can use this information to influence future conservation efforts.
and how hybrid species of salamanders can outcompete other species so well. Looking into the future, the WGER ratio could be compared within an individual (at different environmental conditions), across populations, or even across species to provide insight into breathing efficiency and the relationship between WGER and other metrics of fitness in animal evolution.2 This study provides evidence for hybrid vigor that could have serious implications on the future of species as the climate shifts. More and more hybrids will be created to outcompete their parents, creating high survival rates within new species as the environment changes. However, because hybridization is one of the leading threats to rare species, this may mean native biodiversity will be lost in the future as the climate shifts and species adapt more rapidly.4 Using the WGER ratio, further studies may help us predict the

1. Burger, I. J.; Carter, E. T.; Magner, L. M.; Muñoz, M. M.; Sears, M. W.; Fitzpatrick, B. M.; Riddell, E. A. Functional Ecology 2023.
2. Burger, I. J.; Carter, E. T.; Magner, L. M.; Muñoz, M. M.; Sears, M. W.; Fitzpatrick, B. M.; Riddell, E. A. Functional Ecology: Plain Language Summaries. 2023.
3. Todesco, M.; Pascual, M. A.; Owens, G. L.; Ostevik, K. L.; Moyers, B. T.; Hübner, S.; Heredia, S. M.; Hahn, M. A.; Caseys, C.; Bock, D. G.; Rieseberg, L. H. Hybridization and Extinction. Evolutionary Applications 2016, 9 (7), 892–908.
4. Interview with Dr. Eric Riddell, Ph.D. 02/5/24.

Understanding the ways ecosystems recover from disasters often highlights the important traits that allow life to persevere. Exploring biotic recovery is pivotal for both understanding the past and preparing for the future. In evolutionary biology, biotic recovery refers to the process by which ecosystems recover from disruptions, such as volcanic eruptions. Researchers often observe the stages and pace of biotic recovery by examining species richness and composition at different geological sites. Models and records on previous biotic recovery shed light on new insights and solutions to prevent other potential extinctions from happening. For example, morphological traits that enabled certain species’ survival through mass extinctions might provide ideas for future animal conservation.
Dr. Jordan Claytor, Teaching Assistant Professor of Biology at the University of North Carolina at Chapel Hill, studies mammal species from the early Paleocene and revised a model of biotic recovery from the Cretaceous-Paleogene mass extinction.1 For context, the Paleocene is a geological period lasting from 66–55 million years ago, and the Cretaceous-Paleogene mass extinction also referred to as K-Pg, is a mass extinction that occurred around 66 million years ago. In this study, Dr. Claytor investigates patterns of biotic recovery of mammals through fossils from sites in Montana. His research focuses on three main themes: (1) how certain morphological traits support species to interact better, survive, and reproduce in harsh environments (2) the specific stages of the post-K-Pg biotic recovery of mammals, and (3) the differences between paleontological sites in terms of species diversity. The study’s conclusion proposes a new model of biotic recovery with four stages including the Early Disaster Sub-phase, the Late Disaster Sub-phase, the Early Recovery Subphase, and the Late Recovery Sub-phase. Each
sub-phase includes a different species composition as shown in Figure 2.
The methods used in this study are separated into two parts: a field study and a lab study. The field study spanned six weeks when Dr. Claytor and his team collected fossils from specific layers of Montanan rocks.2 The lab study involved identifying different fossilized species found in those rocks and measuring the fossils’ traits. With this data, Dr. Claytor and his team compared these fossils and others from more dig sites. Through analyzing fossil traits, they were able to compare fossil ages on the geologic timeline to make inferences about species diversity and the process of biotic recovery.
Throughout the study, Dr. Claytor paid special attention to three localities in Montana: Morales 1, Herpijunk Promontory, and Carrie Padgett 6 which all contain different types of rocks, numbers of geographic layers, and species. Montana is a valuable site for archeologists and paleontologists due to its rich prehistoric and historic heritage.2 It is one of the few regions with continuous records of fossils due to a combination of rich geological, climatic, and historical factors. Hence, this
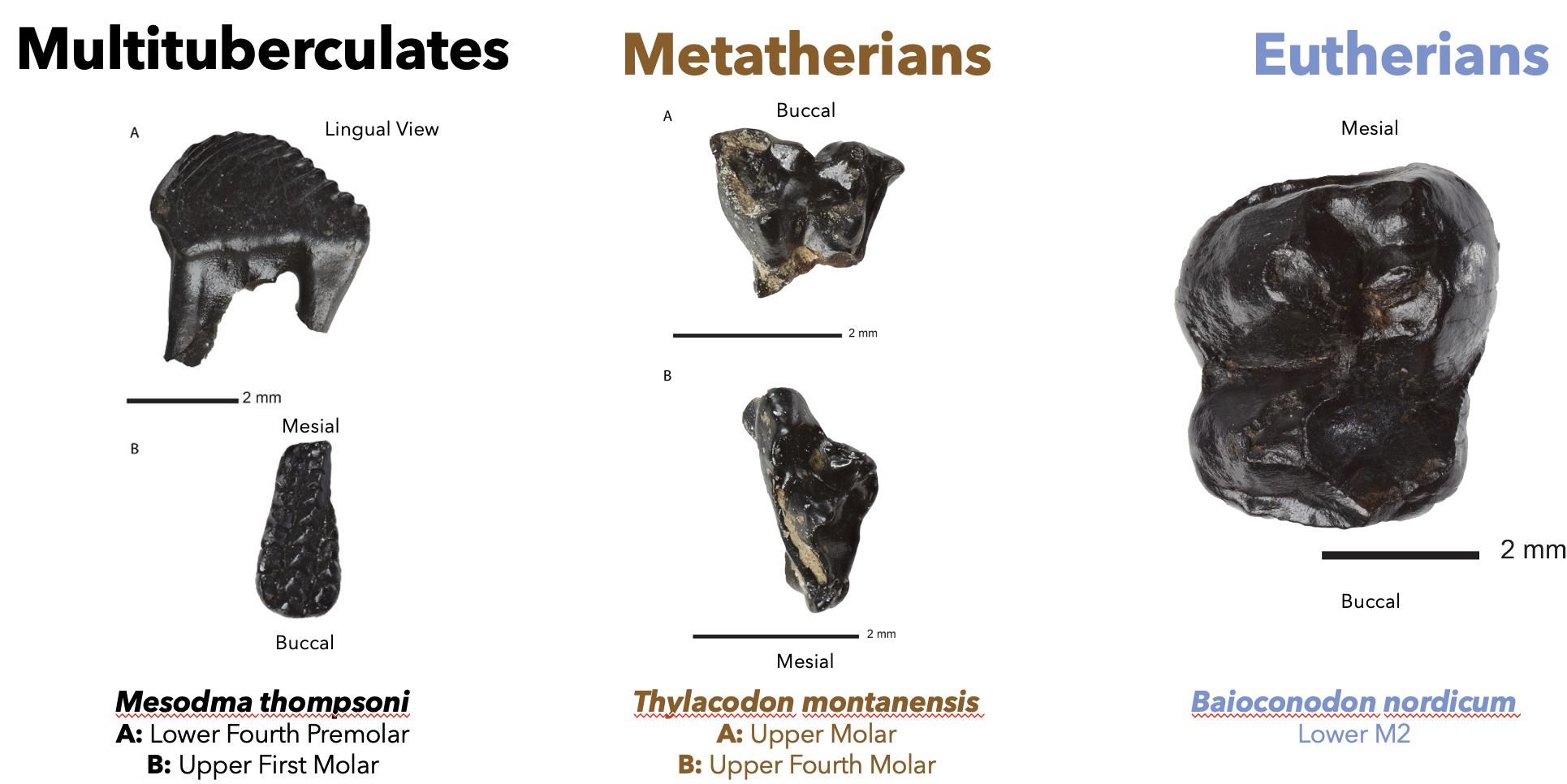
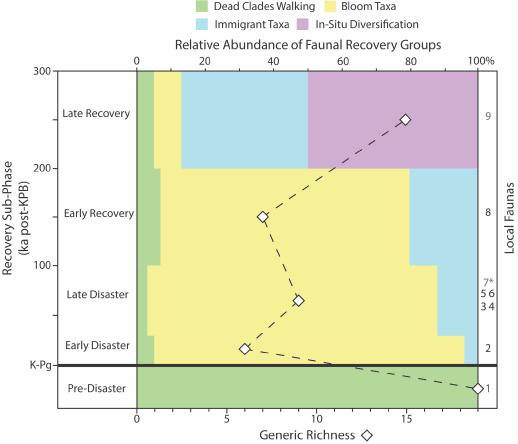
Figure 2. The relative abundance of faunal recovery groups. The X-axis represents the species’s genetic richness, and the Y-axis represents the different stages in the model. The four colors represent different types of species that existed during the time. ‘Dead clades walking” taxa refers to those species that suffered a major decline at a mass extinction but did not completely disappear. Image courtesy of Claytor et al.
area assists researchers like Dr. Claytor in better exploring and understanding the comprehensive evolutionary process. Additionally, Montana contains evidence of diverse prehistoric ecosystems: millions of years ago, plains, mountains, rivers, and other landscapes existed there.4
Most of the lab’s collected fossils were very small mammalian teeth ranging from one millimeter to five millimeters (Figure 1). Dr. Claytor identified different species’ teeth according to their physical traits. The identification of small fossils like these was not easy, especially with many of them being incredibly weathered or damaged.2 While some fossils could not be identified, the team could still draw useful conclusions on the species richness, evenness, and dissimilarity—species richness being the number of species in the ecosystem, and species evenness the measure of how equal the population sizes of different species are within an ecosystem.
The result of the species richness calculation revealed that the Herpijunk locality contains the highest richness value.1 The team calculated Simpson’s and Pielou’s Species Evenness to investigate changes in mammal community structures. While these two metrics involve complex formulas, one could understand them as the distribution and abundance across species within a community.3 A high value of species richness and evenness indicates the locality has a more even distribution of relative abundances across species. The Lancian Flat Creek local fauna, a site studied previously, has high evenness.1 Meanwhile, there is an increase in evenness for both Luck O Hutch and the Littleton local fauna between 100,000 to 300,000 years ago.1 The team used the Bray–Curtis (BC) dissimilarity values to indicate how dissimilar two local faunas are in species composition and relative abundance structure, with higher values indicating a greater degree of dissimilarity between local faunas. Despite the Littleton and Worm Coulee 1 localities, all other localities indicated similar composition and relative abundance of mammals.1
In conclusion, Dr. Claytor’s study proposes a new model of biotic recovery with four stages from the Post-KPB Disaster in northeastern Montana. In the original Three Stages Model, there are only three sub-phases: Early Disaster, Later Disaster, and Late Recovery. Yet, the findings of this study offer new evidence of an additional sub-phase in the biotic recovery model: Early Recovery Sub-phase.1 Through observing species richness from different localities and layers of rocks, researchers identified these four sub-phases in the timeline and the corresponding dominance of different species (Figure 2).1 Dr. Claytor and his team also noticed the difference in the pattern and timing of mammal succession between localities.2 While more studies are needed to examine the accuracy of the model, Dr. Claytor’s research sheds light on how species diversity transformed during ancient times and provides insights for future studies and environment conservation.

1. Claytor, Jordan R., et al. “New Mammalian Local Faunas from the First ca. 80 Ka of the Paleocene in Northeastern Montana and a Revised Model of Biotic Recovery from the Cretaceous–Paleogene Mass Extinction.” Journal of Vertebrate Paleontology, vol. 42, no. 6, Dec. 2022, p. e2222777, https:// doi.org/10.1080/02724634.2023.2222777.
2. Interview with Claytor, Jordan, Ph.D. 02/08/2024
3. Alroy, John. “On Four Measures of Taxonomic Richness.” Paleobiology 46.2 (2020): 158–175. Web.
4. Barnes, B Davis et al. “Dead clades walking are a pervasive macroevolutionary pattern.” Proceedings of the National Academy of Sciences of the United States of America vol. 118,15 (2021): e2019208118. doi:10.1073/pnas.2019208118

Sarah Giang Editor-in-Chief

Meitra Kazemi Managing Editor

Hari Patel Fundraising Chair

Julia Boltz Associate Editor

Isaac Hwang Editor-in-Chief

Ambika Bhatt Treasurer

Sreya Upputuri Online Content Manager

Sprihaa Kolanukuduru Associate Editor

Wan Design Editor

Natalie Druffner Secretary

Esha Agarwal Associate Editor

Andrew Phan Associate Editor

Copy Editor

Heidi Cao Publicity Chair

Kruti Bhargav Associate Editor

Jasmeet Singh Associate Editor
Are you interested in communicating science to a broad audience?
Do you want to engage in thought-provoking invesitgations?
Does your passion for the sciences extend into the world of research?
Do you want to combine your creative talents with your fascination with science? Carolina Scientific is always looking for staff writers, designers, and illustrators! If you are interested, please contact carolina.scientific@gmail.com Follow us on Instagram @carolinascientific Find us on Facebook facebook.com/CarolinaScientific Follow us on Twitter @UNCSci Check out our website carolinascientific.org




“In questions of science, the authority of a thousand is not worth the humble reasoning of a single individual.”
- Galileo Galilei


Spring 2024 | Volume 18 | Issue 2
This publication was funded at least in part by Student Fees which were appropriated and dispersed by the Student Government at UNC-Chapel Hill as well as the Carolina Parents Council.