Statistical issues in survival analysis (Meta-analysis & RMSE)
February 26, 2025
The authors have used network meta-analysis (NMA) to compare multiple treatments simultaneously using restricted mean survival time (RMST) with individual level data (IPD). Since the NMA was mainly used with aggregate data, they had to extend this technique to work with IPD and also RMST. NMA has been an extension of pairwise meta-analysis. They can leverage information about both “direct” and “indirect” comparisons of treatment. There are two stages for the IPD-NMA. The two stage one first estimates treatment effects within each study and then combines study-level effects across studies using NMA while a one stage approached combines the IPD of all studies using a hierarchical model. There are various benefits of using one approach versus the other. In the use of RMST with IPD-NMA, people were using a twostage approach where the RMST is evaluated on IPD in the first stage and then the data is aggregated together in the second stage, but they state this method didn’t properly adjust for covariates in the first stage. Therefore, the authors have tried to develop a one stage IPD-NMA using RMST. They also still proposed advanced two stage methods besides one stage methods for time-to-event outcomes estimated by RMST.
In their description of their two stage RMST IPD-NMA model, it was shown to fit an inverse probability of censoring weighted RMST regression in the first stage also using a log link function. In the second stage, the estimated log RMSTs and treatment by covariate interactions are analyzed using a multivariate meta-analysis model. In their one stage RMST IPD-NMA model, they used a penalized quasi-likelihood (PQL) method to fit the one-stage model with random effects. They implemented the PQL method under the glmmPQL R function available in library MASS. To reduce numerical difficulties, they suggested now using too many covariates and interactions, removing non-existent random effects, and using covariance matrices for random effects with structure like diagonal or blocked diagonal rather than unstructured.
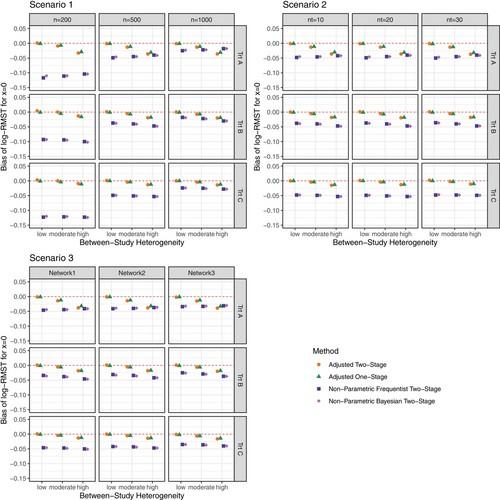
They compared their method to two other two stage models by other researchers, one a frequentist method and the other a Bayesian method. They developed an R package, IPDNMARMST for their proposed and existing method. They ran simulations to compare their two stage and one stage RMST IPD-NMA models under various scenarios and compared to other existing method in terms of bias, mean squared error, and coverage probability of the RMST estimators. Hey varied total sample size of NMA data, number of trials included in NMA, and network of treatments. The biases and mean-squared error decreased with larger sample sizes. With coverage probability, their two stage method showed best coverage than the one stage method, which indicated the standard errors of the log-RMST were underestimated. They concluded their proposed RMST IPD-NMA models outperformed the existing models with smaller biases, lower mean-squared error, and coverage probabilities close to 95% across all scenarios. They in addition verified their methods in a real data example as well.
In their discussion they said that another estimation method besides PQL could have been used here. Another issue they had not addressed was missingness and how that might be handled. Further research is needed in that area.
Written by, Usha Govindarajulu
Keywords: survival analysis, meta-analysis, RMSE, NMA, PQL
References:
Hua K, Wang X, and Hong H (2025). “Network Meta-Analyis of Time-to-Event Endpoints With Individual Participant Data Using Restricted Mean Survival Time Regression” Biometrical Journal. https://doi.org/10.1002/bimj.70037.
https://onlinelibrary.wiley.com/cms/asset/ce1d870d-eccd-4ae9-a1e4-f2ebaf4020e4/bimj70037fig-0001-m.jpg
