12 minute read
Lauren Grice, Chandler Fountain, Michel Modo
Tractography reveals patterns of hippocampal innervation in the human temporal lobe
Lauren Grice c , Chandler Fountain b , and Michel Modo a-c a McGowan Institute for Regenerative Medicine, b Department of Radiology, c Department of Bioengineering, d University of Pittsburgh Medical Center, University of Pittsburgh, Pittsburgh, PA, USA Regenerative Imaging Laboratory, McGowan Institute for Regenerative Medicine, University of Pittsburgh, PA, USA
Lauren Grice
Lauren Grice is a senior Bioengineering major with minors in Electrical Engineering and Neuroscience. She is motivated by understanding the merger between healthcare and technology. After graduation, plans to attend graduate school to study the development of brain computer interfaces for the treatment of neurological disease.
Dr. Michel Modo
Dr. Michel Modo holds a PhD in Neuroscience from King’s College London. He joined the Department of Radiology at the University of Pittsburgh in 2011 and his laboratory’s main focus is to develop novel imaging tools to visualize brain repair using stem cells and biomaterials.
Significance Statement
The temporal lobe is an important brain structure affected by Alzheimer’s disease and epilepsy. This study seeks to establish maps of neuronal connections between grey matter regions in the temporal lobe. Importantly, it is shown that diffusion tensor imaging can effectively characterize grey matter connections in the brain.
Category: Experimental research Keywords: temporal lobe, diffusion tensor imaging, tractography
Abstract
Until recently, medical imaging technology has not been sophisticated enough to develop maps of neuronal connections within the human brain. Such information would be immensely useful as it could help to identify the causes and prognoses of neurological diseases. Diffusion tensor imaging (DTI) is a form of magnetic resonance imaging that can detect the water diffusion vectors in an image. Using tractography, said vectors can be computationally “traced” to create visual representations of neuronal connections. Although DTI has previously been used to identify white matter tracks in the human brain, little research has shown neuronal connectivity within and between grey matter regions. This study applies DTI and tractography to develop a map of hippocampal connections within a healthy human temporal lobe. The results show that the healthy, human hippocampus innervates with two main grey matter structures: the amygdala and the inferior temporal gyrus. This is a significant finding because in the case of disease, changes in the interconnectivity of the hippocampus with the amygdala and inferior temporal gyrus could be an indication of disease onset or prognosis.
1. Introduction
Historically, research on neurological disease has focused on modeling whole brain structures to identify abnormalities. However, because of technological limitations, little research has been performed to establish a map of white matter connections between grey matter structures in the human brain. When considering connectivity within the brain, the temporal lobe (TL) region is of particular interest as it contains important grey matter structures like the hippocampus and amygdala which play significant roles in memory and processing of emotions. Additionally, the TL is targeted by debilitating conditions like epilepsy and Alzheimer’s disease. In an effort to better understand the function of the TL, this project addresses the need for visual maps of microstructural anatomy in a healthy, human TL to develop accurate representations of hippocampal connections and their patterns of cortical innervation.
In this study, mesoscale, T2-weighted, diffusion tensor imaging (DTI) was used to quantify changes in neuronal connectivity. DTI is a specialized form of magnetic resonance imaging (MRI) that is sensitive to the magnitude and orientation of water movement [1]. Software can be used to detect the water diffusion vectors in an image to calculate both the mean diffusivity (MD) and fractional anisotropy (FA), or the directional dependence of diffusion in tissue [2]. Furthermore, using computational methods, the vectors in each voxel, or unit, of a DT image can be weighted by FA and “traced” to create streamlines, or three-dimensional representations of neuronal bundles. In healthy brain tissue, the flow of water molecules in white and grey matter demonstrates low diffusion and tends to be directionally-dependent, or anisotropic. Because of this diffusion behavior, constructed streamlines in DTIs of healthy brains should demonstrate high microstructural organization. However, in conditions like epilepsy and Alzheimer’s disease, brain cells die and
axons degenerate, resulting in altered diffusion behavior and in turn altered streamline organization. Therefore, the knowledge of normal neuronal connections in the TL produced by this study will serve as a valuable point of comparison for future research on underlying structural abnormalities in cases of neurological disease.
2. Methods
The images analyzed in this study were from a whole, postmortem, left human TL. Diffusion weighted MR data were acquired on a 9.6T Bruker BioSpin MRI (DTI EPI; TR = 500 ms; TE = 0.384 ms; b-values = 1000, 2000, 4000, 8000 s/mm2; number of directions per shell = 40, slice thickness = 0.5 mm, FOV = 62.7 mm x 3.8 mm x 97.9 mm; Matrix = 128 x 128 x 196; total number of voxels = 1,218,380; Resolution = 0.01256 mm 3 /voxel). Using DSI Studio, a tractography software, three-dimensional maps of anatomical regions of interest (ROIs) were manually drawn on the MR images showing MD. ROIs delineated grey matter structures of the hippocampus, subiculum, amygdala, caudate, putamen, and TL gyri (i.e. the temporal pole, planum polare, superior temporal gyrus, middle temporal gyrus, inferior temporal gyrus, Heschl’s gyrus, fusiform gyrus, parahippocampal gyrus, planum temporale, transverse gyrus, occipito-temporal gyrus, and lingual gyrus). The ROIs were drawn by selecting appropriate voxels on each slice of the DT image. Then, the selected voxels for each ROI were compiled to create a 3-D representation from which a corresponding FA value and streamline count were extracted. Deterministic tractography was performed to trace streamlines within and extending from the hippocampus.
Tractographic reconstruction parameters included a FA threshold of 0.02, angular threshold of 60°, step size of 0.25 mm (half the size of a voxel edge), minimum length of 1 mm (length of a connection between at least two voxels), and a maximum length of 60 mm (sample height). All seed orientations were considered with sub voxel seed positions and randomized sampling. All fibers passing through, originating, or ending within the seed ROI, or hippocampus, were considered. Streamline direction interpolation was determined using a Euler tracking algorithm terminated after 12,183,800 random seeds for the whole sample (10x the number voxels for total sample). Streamlines were overlaid on an MD map to provide an anatomical reference for their location.
3. Results 3.1 Anatomical verses diffusion images
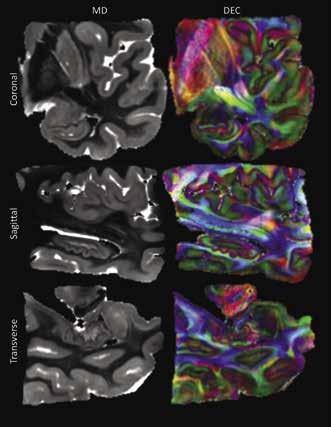
Figure 1 shows three anatomical views of the acquired image from the temporal lobe. The MD images are shown in the left-hand column and highlight a differentiation between white and grey matter, with grey matter regions delineated by a lighter grey color. The right column shows the corresponding diffusion encoded color (DEC) images. These images show a similar delineation between white and grey matter, possibly indicating a discrepancy in diffusion anisotropy between the two tissue subtypes. It is of note the central area of the lobe is shown as mostly blue or green, representing uniform diffusion direction. On the other hand, the cortex and hippocampus regions are multicolored, suggesting isotropic diffusion behavior occurred within the grey matter regions.
Figure 1: The left column shows a coronal, sagittal, and transverse view of the temporal lobe considered in this study. In these images the light grey regions represent areas of higher average diffusion and likely represent grey matter structures such as the hippocampus and cortex. The right column shows the diffusion encoded color images that correspond to the same slices shown in the MD images. Colors in these images group neurons with the same directional orientation.
3.2 Visualization of whole TL streamlines and ROIs
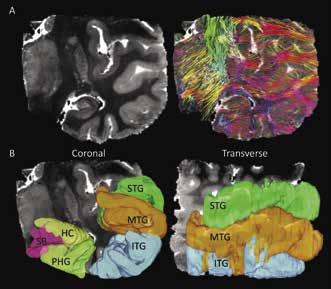
Figure 2A shows a coronal slice of the MD image compared to the same image overlaid with all reconstructed streamlines within it. When comparing the streamlines shown in Figure 2A and the ROIs in Figure 2B, it can be determined that streamlines seem to be less dense in grey matter regions than in white matter regions. Additionally, it is shown that white matter regions appear to be mostly arranged in in the medial/lateral orientation. In Figure 3B at the tail, or fimbria, of the posterior HC, axonal projections join the selenium of the corpus callosum (scc) and a telencephalic white matter tract, the cingulum bundle (cgb). This result verifies the previously known connection of the HC to the cgb and supports the fact that DTI can accurately contribute to the discovery of grey matter interconnectivity within the TL [3].
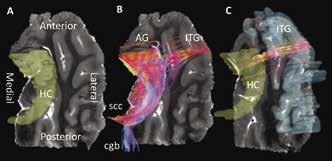
Figure 2: (A) Coronal view of one of the TL MD maps compared to the same image showing all generated streamlines from reconstruction by computational detection and tracing of the FA vector in each voxel in the image. Streamline colors represent its 3D direction in space, with red=lateral/ medial, green=superior/inferior, and blue=anterior/posterior. Intermediate colors represent a combination of two directions. (B) Coronal and transverse views showing the three-dimensional region of interest maps drawn on the DTI image. Here the superior, medial, and inferior temporal gyri are shown (labeled as STG, MTG, and ITG), as well as the parahippocampal gyrus (PHG), subiculum (SB), and hippocampus (HC).
3.3 Hippocampal tractography
Figure 3B illustrates extra- and intrahippocampal streamlines were successfully reconstructed from the hippocampus ROI shown in Figure 3A. Furthermore, this approach demonstrates for the first time that visual representations of both white and grey matter connectivity in the TL can be probed using tractography. The visualization of neuronal connections in Figure 3B and 3C show a precise location for streamline termination as well as provide a qualitative understanding of their arrangement. In Figure 3B, at the anterior end of the HC, efferent neurons connect to two grey matter structures: the AG and ITG. Connections to the AG are few in number, but highly organized. Streamlines extending to the ITG terminate on both medial and lateral edges of an anterior segment of the gyrus. Conversely, as shown in Figure 3C, efferent connections from the ITG appear to fan out within the HC.
Figure 3: (A) Transverse view of one of the TL MD maps with the hippocampus delineated by a 3D ROI. (B) Result of streamline reconstruction by computational detection and tracing of the FA vector in each voxel in the HC ROI. Streamlines within, extending from, and ending in the HC are shown. Streamline colors represent its 3D direction in space, with red=lateral/medial, green=superior/ inferior, and blue=anterior/posterior. Intermediate colors represent a combination of two directions. Tractography reveals four bundles of neurons extending from the HC. At the anterior HC, streamlines connect to the ITG and AG while at the posterior HC, streamlines join the cingulum bundle (cgb) and selenium of the corpus callosum (scc). (C) Isolated streamlines seeded (beginning) in the ITC ROI and ending in the HC ROI.
4. Discussion
The MD images in Figure 1 support the fact that grey matter has less structural organization than white matter and therefore has higher average diffusion [2]. As a result of this increased diffusion, the grey matter regions show up brighter on the MD images in Figure 1. This result validates that diffusion tensor imaging can effectively delineate grey matter regions and can identify the diffusion directions within them. This is further supported by the DEC images in which the central white matter regions are shown in uniform colors suggesting neurons within the white matter branches have similar directional orientation; the cortex and hippocampus on the other hand are shown in multiple colors suggesting there exists a lesser degree of directional orientation of diffusion.
The tractography image in Figure 2A may show a lack of streamline density within grey matter regions because of the limitations of diffusion tensor imaging to differentiate crossing or “kissing” fibers. In other words, although DTI can still identify neuronal bundles in grey matter, it may be limited in its ability to visualize the full complexity of fiber organization within these regions. However, the results in Figure 3 show that tractography was able to reveal that the main grey matter structures the healthy adult HC in this study innervates with are the AG and ITG. This is a significant finding as further identifying isolated
connections between two or more grey matter regions could reveal neuronal circuits that have not been able to be detected before the use of DTI. Furthermore, previous studies have shown that functions such as perception, imagination, and episodic memory engage the anterior HC [4]. Since the HC plays a major role in memory formation and has been shown to atrophy in cases of Alzheimer’s disease and dementia, it is possible that a change in the interconnectivity of the HC with the AG and ITG could be an indication of disease prognosis [5]. Furthermore, previous studies have theorized that loss of connections precedes atrophy in Huntington’s disease. If this degeneration paradigm is consistent in Alzheimer’s disease, it may be possible to use DTI to identify the onset of dementia before the patient begins to show severe symptoms [6].
5. Conclusion
The methods in this study revealed that diffusion tensor imaging, although mainly used to identify white matter tracks in previous studies, can successfully reconstruct streamlines within grey matter regions as well as reveal patterns of grey matter region interconnectivity. Specifically, the results of this study showed that the main grey matter structures a healthy adult HC innervates with are the AG and ITG. Chiefly, though, the significance of reconstructing streamlines from DTIs in this study was to gain an accurate, comprehensive, and novel characterization of tissue microstructure within the TL and, specifically, the HC. Importantly, reconstructed streamlines from this study will later help to characterize the pathological basis of diseases that manifest in the TL, like Alzheimer’s disease.
6. Acknowledgements
Lauren Grice was supported by the University of Pittsburgh Swanson School of Engineering and Office of the Provost throughout the entirety of this research.
7. References
[1] Mori & Zhang. “Principles of diffusion tensor imaging and its application to basic neuroscience research.” Neuron 51(5), 527- 539, 2006, doi: 10.1016/j.neuron.2006.08.012.
[2] C.H. Sotak. “The role of diffusion tensor imaging in the evaluation of ischemic brain injury – a review.” NMR in Biomedicine 15(7-8), 561-569, 2002, doi: 10.1002/nbm.786.
[3] Wu et al. “Segmentation of the Cingulum Bundle in the Human Brain: A New Perspective Based on DSI Tractography and Fiber Dissection Study.” Frontiers in Neuroanatomy 10, 10-84, 2016, doi: 10.3389/fnana.2016.00084.
[4] Zeidman & Maguire. “Anterior hippocampus: the anatomy of perception, imagination and episodic memory.” Nature Reviews Neuroscience 17(3), 173–182, 2016, doi: 10.1038/nrn.2015.24. [5] Mu & Gage. “Adult hippocampal neurogenesis and its role in Alzheimer’s disease.” Molecular Neurodegeneration, 6, 6-85, 2011, doi: 10.1186/1750-1326-6-85.
[6] McColan et al. ”Brain Regions Showing White Matter Loss in Huntington’s Disease Are Enriched for Synaptic and Metabolic Genes” Biological Psychiatry 83(5), 456-465, 2018, doi: 10.1016/j.biopsych.2017.10.019.










